Gene
KWMTBOMO07508
Annotation
UDP-glycosyltransferase_UGT46B1_[Helicoverpa_armigera]
Full name
UDP-glucuronosyltransferase
Location in the cell
Extracellular Reliability : 1.118
Sequence
CDS
ATGAAAGCCATAATCAACGCTTTTGCTCGTTTACCACAAACAATATTTTGGAAATGGGAGGACGATATCTCAGACGCGATAAAAATACCAAAGAACGTTAAATTTGAAAAATGGTTGCCGCAATACGATTTGTTGAACCAGCAAAATTGCTTGGCATTTATCACACACGGCGGTCTACTTAGCTTGACGGAGGCAGTCGCGTCAAGCGTGCCGACTCTCATTATTCCTATACTCGGAGATCAGTTCGGGAACGCGGCCCATGCTCAGAAAGCTGGCGTCGCCGAGGTTCTAAGTCTTACGGAAATTGAAGAAGACGTGTTCTATAATAAACTCTTATCAGTTTTGTCTAAGGAGAAACGTCAGAAAGCTAAATTACTAACAAGTCTTTGGCGGGATCGTCCTCAACCGGCAATGGAAACGGCCCTATTCAACATAGAACTCGTAGCGCGACATAAAGGACTGGATATGTCGTCTGACCTATATTTCCGTAATTTATTGTTACCCACCTGTTTTGGGAATTCGTTAATAGCCATGAGCACTGCTAAGCTAATGCTCATAGCTGGAGAGCTTAAGAGGAATCAGCAAAGAGTTCCAGTACAAAACTACTACTAA
Protein
MKAIINAFARLPQTIFWKWEDDISDAIKIPKNVKFEKWLPQYDLLNQQNCLAFITHGGLLSLTEAVASSVPTLIIPILGDQFGNAAHAQKAGVAEVLSLTEIEEDVFYNKLLSVLSKEKRQKAKLLTSLWRDRPQPAMETALFNIELVARHKGLDMSSDLYFRNLLLPTCFGNSLIAMSTAKLMLIAGELKRNQQRVPVQNYY
Summary
Catalytic Activity
glucuronate acceptor + UDP-alpha-D-glucuronate = acceptor beta-D-glucuronoside + H(+) + UDP
Similarity
Belongs to the UDP-glycosyltransferase family.
Feature
chain UDP-glucuronosyltransferase
Uniprot
A0A2H1VU07
G9LPR8
A0A2A4JI28
V5H2B2
A0A2H8TIZ0
A0A1Y1LMI4
+ More
A0A1Y1LL43 A0A1Y1LIT5 A0A2H4WB74 A0A1L8D6F8 A0A1W4WYE4 A0A1W4WMF1 A0A1W4WMC9 D2A2V5 A0A286MXP5 A0A1W4WY61 A0A139WIW7 A0A212EWF8 S4PCR1 A0A191T1N3 A0A286MXP6 G9LPP9 G9LPR5 A0A2S2PC03 A0A2H8TVS6 A0A2D1GSK6 V9IL65 A0A2A4JHL4 J9KB24 A0A1W7R8H2 J9K7F3 A0A182GYX7 A0A2D1GSG4 A0A1B6KVV9 A0A2H1W091 A0A3S2PBM0 A0A2D1GSL9 A0A1B6L8F2 A0A1B6FW51 A0A194PN21 A0A1B6KXR8 A0A1B6F5J1 A0A2R4FXJ3 Q17KZ7 A0A2H1W5A1 A0A2S2P7P3 G9LPV9 A0A2S2NP66 A0A2A4K0B3 A0A1S4EZ13 G9LPV6 A0A0L7L3K3 N6UB74 A0A087ZYR9 J3JVI3 A0A191T1L9 A0A2D1GSI9 A0A0P5JI56 A0A2D1GSF6 A0A2J7R4T0 A0A2P8ZG78 A0A3S2N8S7 A0A336LTZ6 A0A2H8TXR1 A0A026WYL1 A0A3L8DPI9 A0A2S2R018 A0A2D1GSM0 A0A1D2MYE5 A0A1S3CYB2 A0A194PHJ2 X1WIH9 A0A0P5D4L6 A0A162P1L4 A0A1D2NEE8 A0A0P6E0N5 A0A0N8CHL9 A0A194PIG8 A0A2P8ZG70 E9HIB8 V5GU86 A0A0P5VPG3 A0A2J7R4W0 A0A0N8DAL2 A0A0L7RH14 A0A226DZS8 A0A023R9H7 A0A2H1VD70 A0A067RAP7 B0VZV7 A0A023R6Z5 A0A2H8TRY0 A0A2J7R4W5 A0A191T1P6 G9LPR3 N6UH99 A0A1B6E1B5 A0A1B6DPF5 A0A1B6DAQ2 A0A1B6CCZ4
A0A1Y1LL43 A0A1Y1LIT5 A0A2H4WB74 A0A1L8D6F8 A0A1W4WYE4 A0A1W4WMF1 A0A1W4WMC9 D2A2V5 A0A286MXP5 A0A1W4WY61 A0A139WIW7 A0A212EWF8 S4PCR1 A0A191T1N3 A0A286MXP6 G9LPP9 G9LPR5 A0A2S2PC03 A0A2H8TVS6 A0A2D1GSK6 V9IL65 A0A2A4JHL4 J9KB24 A0A1W7R8H2 J9K7F3 A0A182GYX7 A0A2D1GSG4 A0A1B6KVV9 A0A2H1W091 A0A3S2PBM0 A0A2D1GSL9 A0A1B6L8F2 A0A1B6FW51 A0A194PN21 A0A1B6KXR8 A0A1B6F5J1 A0A2R4FXJ3 Q17KZ7 A0A2H1W5A1 A0A2S2P7P3 G9LPV9 A0A2S2NP66 A0A2A4K0B3 A0A1S4EZ13 G9LPV6 A0A0L7L3K3 N6UB74 A0A087ZYR9 J3JVI3 A0A191T1L9 A0A2D1GSI9 A0A0P5JI56 A0A2D1GSF6 A0A2J7R4T0 A0A2P8ZG78 A0A3S2N8S7 A0A336LTZ6 A0A2H8TXR1 A0A026WYL1 A0A3L8DPI9 A0A2S2R018 A0A2D1GSM0 A0A1D2MYE5 A0A1S3CYB2 A0A194PHJ2 X1WIH9 A0A0P5D4L6 A0A162P1L4 A0A1D2NEE8 A0A0P6E0N5 A0A0N8CHL9 A0A194PIG8 A0A2P8ZG70 E9HIB8 V5GU86 A0A0P5VPG3 A0A2J7R4W0 A0A0N8DAL2 A0A0L7RH14 A0A226DZS8 A0A023R9H7 A0A2H1VD70 A0A067RAP7 B0VZV7 A0A023R6Z5 A0A2H8TRY0 A0A2J7R4W5 A0A191T1P6 G9LPR3 N6UH99 A0A1B6E1B5 A0A1B6DPF5 A0A1B6DAQ2 A0A1B6CCZ4
EC Number
2.4.1.17
Pubmed
EMBL
ODYU01004162
SOQ43734.1
JQ070225
AEW43141.1
NWSH01001496
PCG71080.1
+ More
GALX01001513 JAB66953.1 GFXV01002302 MBW14107.1 GEZM01054554 JAV73560.1 GEZM01054556 JAV73558.1 GEZM01054555 JAV73559.1 KY202936 AUC64280.1 GEYN01000098 JAV02031.1 KQ971338 EFA02977.2 MF684363 ASX93997.1 KYB27856.1 AGBW02012000 OWR45816.1 GAIX01004036 JAA88524.1 KU680308 ANI22014.1 MF684364 ASX93998.1 JQ070206 AEW43122.1 JQ070222 AEW43138.1 GGMR01014372 MBY26991.1 GFXV01005857 MBW17662.1 MF471430 ATN96036.1 JR050268 AEY61216.1 NWSH01001552 PCG70882.1 ABLF02020518 ABLF02020523 ABLF02020526 GEHC01000199 JAV47446.1 ABLF02020524 ABLF02020525 JXUM01021599 KQ560606 KXJ81644.1 MF471406 ATN96012.1 GEBQ01024407 JAT15570.1 ODYU01005418 SOQ46242.1 RSAL01000119 RVE46733.1 MF471452 ATN96058.1 GEBQ01020086 JAT19891.1 GECZ01015337 JAS54432.1 KQ459604 KPI92520.1 GEBQ01023933 JAT16044.1 GECZ01024322 GECZ01008492 JAS45447.1 JAS61277.1 MF034860 AVT42226.1 CH477220 EAT47358.1 ODYU01006427 SOQ48259.1 GGMR01012806 MBY25425.1 JQ070266 AEW43182.1 GGMR01005927 MBY18546.1 NWSH01000322 PCG77489.1 JQ070263 AEW43179.1 JTDY01003254 KOB69869.1 APGK01030497 APGK01030498 KB740774 ENN78970.1 BT127251 AEE62213.1 KU680293 ANI21999.1 MF471426 ATN96032.1 GDIQ01202544 JAK49181.1 MF471427 ATN96033.1 NEVH01007402 PNF35837.1 PYGN01000068 PSN55501.1 RSAL01000196 RVE44553.1 UFQT01000109 SSX20153.1 GFXV01007278 MBW19083.1 KK107063 EZA61120.1 QOIP01000005 RLU22281.1 GGMS01013519 MBY82722.1 MF471451 ATN96057.1 LJIJ01000389 ODM98012.1 KPI92518.1 ABLF02037495 ABLF02037496 ABLF02037503 GDIP01162987 JAJ60415.1 LRGB01000512 KZS18442.1 LJIJ01000067 ODN03640.1 GDIQ01074138 JAN20599.1 GDIP01130895 JAL72819.1 KPI92519.1 PSN55494.1 GL732654 EFX68472.1 GALX01004658 JAB63808.1 GDIP01097242 JAM06473.1 PNF35865.1 GDIP01052551 JAM51164.1 KQ414596 KOC70104.1 LNIX01000009 OXA50201.1 KJ584814 AHX56940.1 ODYU01001909 SOQ38775.1 KK852590 KDR20745.1 DS231815 EDS35199.1 KJ584823 AHX56948.1 GFXV01004547 MBW16352.1 PNF35871.1 KU680306 ANI22012.1 JQ070220 AEW43136.1 APGK01025969 KB740608 ENN80011.1 GEDC01005597 JAS31701.1 GEDC01009759 JAS27539.1 GEDC01014521 JAS22777.1 GEDC01025996 JAS11302.1
GALX01001513 JAB66953.1 GFXV01002302 MBW14107.1 GEZM01054554 JAV73560.1 GEZM01054556 JAV73558.1 GEZM01054555 JAV73559.1 KY202936 AUC64280.1 GEYN01000098 JAV02031.1 KQ971338 EFA02977.2 MF684363 ASX93997.1 KYB27856.1 AGBW02012000 OWR45816.1 GAIX01004036 JAA88524.1 KU680308 ANI22014.1 MF684364 ASX93998.1 JQ070206 AEW43122.1 JQ070222 AEW43138.1 GGMR01014372 MBY26991.1 GFXV01005857 MBW17662.1 MF471430 ATN96036.1 JR050268 AEY61216.1 NWSH01001552 PCG70882.1 ABLF02020518 ABLF02020523 ABLF02020526 GEHC01000199 JAV47446.1 ABLF02020524 ABLF02020525 JXUM01021599 KQ560606 KXJ81644.1 MF471406 ATN96012.1 GEBQ01024407 JAT15570.1 ODYU01005418 SOQ46242.1 RSAL01000119 RVE46733.1 MF471452 ATN96058.1 GEBQ01020086 JAT19891.1 GECZ01015337 JAS54432.1 KQ459604 KPI92520.1 GEBQ01023933 JAT16044.1 GECZ01024322 GECZ01008492 JAS45447.1 JAS61277.1 MF034860 AVT42226.1 CH477220 EAT47358.1 ODYU01006427 SOQ48259.1 GGMR01012806 MBY25425.1 JQ070266 AEW43182.1 GGMR01005927 MBY18546.1 NWSH01000322 PCG77489.1 JQ070263 AEW43179.1 JTDY01003254 KOB69869.1 APGK01030497 APGK01030498 KB740774 ENN78970.1 BT127251 AEE62213.1 KU680293 ANI21999.1 MF471426 ATN96032.1 GDIQ01202544 JAK49181.1 MF471427 ATN96033.1 NEVH01007402 PNF35837.1 PYGN01000068 PSN55501.1 RSAL01000196 RVE44553.1 UFQT01000109 SSX20153.1 GFXV01007278 MBW19083.1 KK107063 EZA61120.1 QOIP01000005 RLU22281.1 GGMS01013519 MBY82722.1 MF471451 ATN96057.1 LJIJ01000389 ODM98012.1 KPI92518.1 ABLF02037495 ABLF02037496 ABLF02037503 GDIP01162987 JAJ60415.1 LRGB01000512 KZS18442.1 LJIJ01000067 ODN03640.1 GDIQ01074138 JAN20599.1 GDIP01130895 JAL72819.1 KPI92519.1 PSN55494.1 GL732654 EFX68472.1 GALX01004658 JAB63808.1 GDIP01097242 JAM06473.1 PNF35865.1 GDIP01052551 JAM51164.1 KQ414596 KOC70104.1 LNIX01000009 OXA50201.1 KJ584814 AHX56940.1 ODYU01001909 SOQ38775.1 KK852590 KDR20745.1 DS231815 EDS35199.1 KJ584823 AHX56948.1 GFXV01004547 MBW16352.1 PNF35871.1 KU680306 ANI22012.1 JQ070220 AEW43136.1 APGK01025969 KB740608 ENN80011.1 GEDC01005597 JAS31701.1 GEDC01009759 JAS27539.1 GEDC01014521 JAS22777.1 GEDC01025996 JAS11302.1
Proteomes
UP000218220
UP000192223
UP000007266
UP000007151
UP000007819
UP000069940
+ More
UP000249989 UP000283053 UP000053268 UP000008820 UP000037510 UP000019118 UP000005203 UP000235965 UP000245037 UP000053097 UP000279307 UP000094527 UP000079169 UP000076858 UP000000305 UP000053825 UP000198287 UP000027135 UP000002320
UP000249989 UP000283053 UP000053268 UP000008820 UP000037510 UP000019118 UP000005203 UP000235965 UP000245037 UP000053097 UP000279307 UP000094527 UP000079169 UP000076858 UP000000305 UP000053825 UP000198287 UP000027135 UP000002320
Pfam
PF00201 UDPGT
ProteinModelPortal
A0A2H1VU07
G9LPR8
A0A2A4JI28
V5H2B2
A0A2H8TIZ0
A0A1Y1LMI4
+ More
A0A1Y1LL43 A0A1Y1LIT5 A0A2H4WB74 A0A1L8D6F8 A0A1W4WYE4 A0A1W4WMF1 A0A1W4WMC9 D2A2V5 A0A286MXP5 A0A1W4WY61 A0A139WIW7 A0A212EWF8 S4PCR1 A0A191T1N3 A0A286MXP6 G9LPP9 G9LPR5 A0A2S2PC03 A0A2H8TVS6 A0A2D1GSK6 V9IL65 A0A2A4JHL4 J9KB24 A0A1W7R8H2 J9K7F3 A0A182GYX7 A0A2D1GSG4 A0A1B6KVV9 A0A2H1W091 A0A3S2PBM0 A0A2D1GSL9 A0A1B6L8F2 A0A1B6FW51 A0A194PN21 A0A1B6KXR8 A0A1B6F5J1 A0A2R4FXJ3 Q17KZ7 A0A2H1W5A1 A0A2S2P7P3 G9LPV9 A0A2S2NP66 A0A2A4K0B3 A0A1S4EZ13 G9LPV6 A0A0L7L3K3 N6UB74 A0A087ZYR9 J3JVI3 A0A191T1L9 A0A2D1GSI9 A0A0P5JI56 A0A2D1GSF6 A0A2J7R4T0 A0A2P8ZG78 A0A3S2N8S7 A0A336LTZ6 A0A2H8TXR1 A0A026WYL1 A0A3L8DPI9 A0A2S2R018 A0A2D1GSM0 A0A1D2MYE5 A0A1S3CYB2 A0A194PHJ2 X1WIH9 A0A0P5D4L6 A0A162P1L4 A0A1D2NEE8 A0A0P6E0N5 A0A0N8CHL9 A0A194PIG8 A0A2P8ZG70 E9HIB8 V5GU86 A0A0P5VPG3 A0A2J7R4W0 A0A0N8DAL2 A0A0L7RH14 A0A226DZS8 A0A023R9H7 A0A2H1VD70 A0A067RAP7 B0VZV7 A0A023R6Z5 A0A2H8TRY0 A0A2J7R4W5 A0A191T1P6 G9LPR3 N6UH99 A0A1B6E1B5 A0A1B6DPF5 A0A1B6DAQ2 A0A1B6CCZ4
A0A1Y1LL43 A0A1Y1LIT5 A0A2H4WB74 A0A1L8D6F8 A0A1W4WYE4 A0A1W4WMF1 A0A1W4WMC9 D2A2V5 A0A286MXP5 A0A1W4WY61 A0A139WIW7 A0A212EWF8 S4PCR1 A0A191T1N3 A0A286MXP6 G9LPP9 G9LPR5 A0A2S2PC03 A0A2H8TVS6 A0A2D1GSK6 V9IL65 A0A2A4JHL4 J9KB24 A0A1W7R8H2 J9K7F3 A0A182GYX7 A0A2D1GSG4 A0A1B6KVV9 A0A2H1W091 A0A3S2PBM0 A0A2D1GSL9 A0A1B6L8F2 A0A1B6FW51 A0A194PN21 A0A1B6KXR8 A0A1B6F5J1 A0A2R4FXJ3 Q17KZ7 A0A2H1W5A1 A0A2S2P7P3 G9LPV9 A0A2S2NP66 A0A2A4K0B3 A0A1S4EZ13 G9LPV6 A0A0L7L3K3 N6UB74 A0A087ZYR9 J3JVI3 A0A191T1L9 A0A2D1GSI9 A0A0P5JI56 A0A2D1GSF6 A0A2J7R4T0 A0A2P8ZG78 A0A3S2N8S7 A0A336LTZ6 A0A2H8TXR1 A0A026WYL1 A0A3L8DPI9 A0A2S2R018 A0A2D1GSM0 A0A1D2MYE5 A0A1S3CYB2 A0A194PHJ2 X1WIH9 A0A0P5D4L6 A0A162P1L4 A0A1D2NEE8 A0A0P6E0N5 A0A0N8CHL9 A0A194PIG8 A0A2P8ZG70 E9HIB8 V5GU86 A0A0P5VPG3 A0A2J7R4W0 A0A0N8DAL2 A0A0L7RH14 A0A226DZS8 A0A023R9H7 A0A2H1VD70 A0A067RAP7 B0VZV7 A0A023R6Z5 A0A2H8TRY0 A0A2J7R4W5 A0A191T1P6 G9LPR3 N6UH99 A0A1B6E1B5 A0A1B6DPF5 A0A1B6DAQ2 A0A1B6CCZ4
PDB
2O6L
E-value=1.32845e-13,
Score=181
Ontologies
PATHWAY
00040
Pentose and glucuronate interconversions - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00830 Retinol metabolism - Bombyx mori (domestic silkworm)
00860 Porphyrin and chlorophyll metabolism - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00830 Retinol metabolism - Bombyx mori (domestic silkworm)
00860 Porphyrin and chlorophyll metabolism - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
Topology
Subcellular location
Membrane
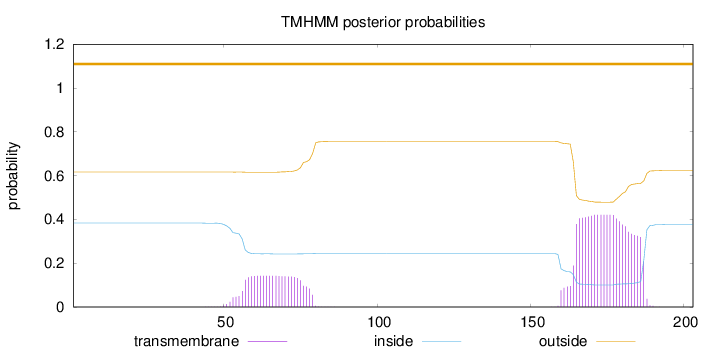
Length:
203
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.59283
Exp number, first 60 AAs:
0.80132
Total prob of N-in:
0.38321
outside
1 - 203
Population Genetic Test Statistics
Pi
2.558615
Theta
5.36592
Tajima's D
-1.986158
CLR
14.853209
CSRT
0.0105494725263737
Interpretation
Uncertain
