Pre Gene Modal
BGIBMGA005878
Annotation
PREDICTED:_transcription_initiation_factor_IIE_subunit_beta_[Papilio_xuthus]
Full name
Transcription initiation factor IIE subunit beta
Location in the cell
Nuclear Reliability : 4.566
Sequence
CDS
ATGGACCCGGCGCTCTTACGGGAACGGGAAGCGTTTAAAAAGAAGGCACTTGCGACCCCGAGCATTGAAAAAAAGAAAAGGGATGACTCTGAATTCAAAGATGACAGCAAAAAGAAATCGAAATCATCCAGCTCAGTCAGCTCCGGACCAAAGCTTGATGCTAGTAATTACAAGACTATGACTGGCAGCTCGTCCTATCGCTTTGGTGTGCTAGCGCGTATTGTACGGCATATGAGAGCAAGGCATCAAGATGGAGATGACCATCCACTGTCTTTAGATGAGATTTTAGATGAGACAAATCAATTGGATGTCGGAAATAAAATAAAACAGTGGCTACAAACAGAAGCCCTACAGAGCAATCCTAAAATCGAATGCTCTCCGGATGGAAAGTTTAACTTCAAACCGGTTTATAAAATAAAAGATAAAAAATCTTTACTGAGGTTACTTAAGCAACAAGACCTCAAAGGCTTGGGAGGTATATTATTGGAAGATGTTCAAGAGTCGCTGCCTCACTGCGAGAGAGCACTGAAACATTTGGCACAAGAGATTTTGTATATAACAAGGCACACTGATAAAAAGAAAATATTATTCTACAATGATAAGACTGCTACATTAGACGTAGATGACGAATTCGTGAAATTATGGCGTGCAACAGCGGTGGACGCGATGGACGACGCGAAGATCGAGGAGTACCTCGAGAAGCAAGGGATCAAATCCATGCAGGATCACGGGCCAAAGAAGATGCTTGCTCCCAAACGGAAGAAGGCCAAGCAGAAGCGTAGACAGTTCAAGAAACCAAGAGACAATGAACATTTGGCCCACGTACTGGAGACGTACGAGGATAACACGCTGACTCAGCAAGGAGTTTCCATTAAATAA
Protein
MDPALLREREAFKKKALATPSIEKKKRDDSEFKDDSKKKSKSSSSVSSGPKLDASNYKTMTGSSSYRFGVLARIVRHMRARHQDGDDHPLSLDEILDETNQLDVGNKIKQWLQTEALQSNPKIECSPDGKFNFKPVYKIKDKKSLLRLLKQQDLKGLGGILLEDVQESLPHCERALKHLAQEILYITRHTDKKKILFYNDKTATLDVDDEFVKLWRATAVDAMDDAKIEEYLEKQGIKSMQDHGPKKMLAPKRKKAKQKRRQFKKPRDNEHLAHVLETYEDNTLTQQGVSIK
Summary
Description
Recruits TFIIH to the initiation complex and stimulates the RNA polymerase II C-terminal domain kinase and DNA-dependent ATPase activities of TFIIH. Both TFIIH and TFIIE are required for promoter clearance by RNA polymerase.
Subunit
Tetramer of two alpha and two beta chains.
Similarity
Belongs to the TFIIE beta subunit family.
Uniprot
H9J8N6
A0A2A4JI13
A0A1E1WHF1
A0A0N1IFX0
S4PY18
A0A2H1V2R2
+ More
A0A212EK30 A0A194PU71 U5EUM5 B0X433 A0A1Q3FA59 A0A0A1XM27 B4L9A3 B4LBQ9 A0A182KW59 A0A182XCZ9 Q7Q7C7 A0A182IKD0 A0A0K8W5D1 T1PH62 W8BZ93 A0A1L8DH07 A0A182UXT4 A0A182HMC7 A0A182R2N9 A0A182SB97 A0A182LZD4 A0A182QG40 A0A182N7T1 A0A034WLD0 A0A084WEU4 A0A182YNP2 W5JAL9 A0A0L0CLB8 A0A023EMR2 A0A182KAI9 A0A182WAN9 A0A2M3Z8F2 A0A2M4AQS9 A0A182FFQ2 A0A2M4BVN0 A0A182H2L8 A0A1I8Q164 A0A1W4VUN4 B4J1I0 A0A182GLZ8 Q29EG4 B4PHT7 A0A182PUU5 B4H1Z4 J3JWI6 B3M909 A0A3B0J9U2 A0A0K8TR33 B4NLY6 A0A1S4FTA4 Q16Q16 B3NC19 O96881 B4HU43 B4QQJ7 D6WSM2 D3TNI3 A0A2H1VAM8 A0A0L7LBA6 A0A1Y1N4G3 A0A2A3EJN2 A0A1B0AR63 A0A2J7QNS9 A0A1A9XGS6 A0A154P2S5 A0A0M4EYZ6 F4WUG3 A0A151XFP8 A0A195BDD9 A0A158NFE3 A0A151JZH5 A0A2P8YW15 A0A195D6U3 A0A0L7R9I5 A0A336LHH7 E2BF31 A0A0J7KPL3 E0VXJ2 E9IJ90 E2A6B3 A0A087ZQE1 A0A1B0CBT6 A0A026WHK3 A0A1B6GPV1 A0A232FF13 A0A1B6HZY4 K7JAG0 A0A1B6DGD1 A0A0A9Y5A9 A0A1B0FRJ7 A0A069DYD8 A0A182UI79 A0A0N0BEF6 A0A1A9ZG27
A0A212EK30 A0A194PU71 U5EUM5 B0X433 A0A1Q3FA59 A0A0A1XM27 B4L9A3 B4LBQ9 A0A182KW59 A0A182XCZ9 Q7Q7C7 A0A182IKD0 A0A0K8W5D1 T1PH62 W8BZ93 A0A1L8DH07 A0A182UXT4 A0A182HMC7 A0A182R2N9 A0A182SB97 A0A182LZD4 A0A182QG40 A0A182N7T1 A0A034WLD0 A0A084WEU4 A0A182YNP2 W5JAL9 A0A0L0CLB8 A0A023EMR2 A0A182KAI9 A0A182WAN9 A0A2M3Z8F2 A0A2M4AQS9 A0A182FFQ2 A0A2M4BVN0 A0A182H2L8 A0A1I8Q164 A0A1W4VUN4 B4J1I0 A0A182GLZ8 Q29EG4 B4PHT7 A0A182PUU5 B4H1Z4 J3JWI6 B3M909 A0A3B0J9U2 A0A0K8TR33 B4NLY6 A0A1S4FTA4 Q16Q16 B3NC19 O96881 B4HU43 B4QQJ7 D6WSM2 D3TNI3 A0A2H1VAM8 A0A0L7LBA6 A0A1Y1N4G3 A0A2A3EJN2 A0A1B0AR63 A0A2J7QNS9 A0A1A9XGS6 A0A154P2S5 A0A0M4EYZ6 F4WUG3 A0A151XFP8 A0A195BDD9 A0A158NFE3 A0A151JZH5 A0A2P8YW15 A0A195D6U3 A0A0L7R9I5 A0A336LHH7 E2BF31 A0A0J7KPL3 E0VXJ2 E9IJ90 E2A6B3 A0A087ZQE1 A0A1B0CBT6 A0A026WHK3 A0A1B6GPV1 A0A232FF13 A0A1B6HZY4 K7JAG0 A0A1B6DGD1 A0A0A9Y5A9 A0A1B0FRJ7 A0A069DYD8 A0A182UI79 A0A0N0BEF6 A0A1A9ZG27
Pubmed
19121390
26354079
23622113
22118469
25830018
17994087
+ More
20966253 12364791 14747013 17210077 25315136 24495485 25348373 24438588 25244985 20920257 23761445 26108605 24945155 26483478 15632085 17550304 22516182 23537049 26369729 17510324 9012800 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 18362917 19820115 20353571 26227816 28004739 21719571 21347285 29403074 20798317 20566863 21282665 24508170 30249741 28648823 20075255 25401762 26823975 26334808
20966253 12364791 14747013 17210077 25315136 24495485 25348373 24438588 25244985 20920257 23761445 26108605 24945155 26483478 15632085 17550304 22516182 23537049 26369729 17510324 9012800 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 18362917 19820115 20353571 26227816 28004739 21719571 21347285 29403074 20798317 20566863 21282665 24508170 30249741 28648823 20075255 25401762 26823975 26334808
EMBL
BABH01030376
NWSH01001496
PCG71073.1
GDQN01004718
JAT86336.1
KQ460954
+ More
KPJ10250.1 GAIX01004063 JAA88497.1 ODYU01000407 SOQ35135.1 AGBW02014349 OWR41830.1 KQ459592 KPI96867.1 GANO01001402 JAB58469.1 DS232327 EDS40110.1 GFDL01010605 JAV24440.1 GBXI01002377 JAD11915.1 CH933816 EDW17278.1 CH940647 EDW68686.1 AAAB01008960 EAA11794.3 GDHF01005901 JAI46413.1 KA648111 AFP62740.1 GAMC01001923 JAC04633.1 GFDF01008417 JAV05667.1 APCN01004405 AXCM01002989 AXCN02000586 GAKP01004374 JAC54578.1 ATLV01023258 KE525341 KFB48738.1 ADMH02001932 ETN60443.1 JRES01000240 KNC33041.1 GAPW01002956 JAC10642.1 GGFM01004053 MBW24804.1 GGFK01009826 MBW43147.1 GGFJ01007911 MBW57052.1 JXUM01105523 KQ564973 KXJ71485.1 CH916366 EDV95871.1 JXUM01073321 KQ562767 KXJ75165.1 CH379070 EAL30097.1 CM000159 EDW93396.1 CH479203 EDW30346.1 APGK01010618 APGK01017343 BT127604 KB740012 KB738240 KB631654 AEE62566.1 ENN81901.1 ENN82710.1 ERL85053.1 CH902618 EDV38953.1 OUUW01000002 SPP77163.1 GDAI01000766 JAI16837.1 CH964278 EDW85354.1 CH477766 EAT36460.1 CH954178 EDV50907.1 U72893 AE014296 BT022197 AAD00188.1 AAF47880.1 AAY51591.1 CH480817 EDW50464.1 CM000363 CM002912 EDX09203.1 KMY97561.1 KQ971354 EFA05897.1 EZ422985 ADD19261.1 ODYU01001544 SOQ37910.1 JTDY01001830 KOB72772.1 GEZM01013039 JAV92801.1 KZ288223 PBC32023.1 JXJN01002265 NEVH01012562 PNF30239.1 KQ434795 KZC05528.1 CP012525 ALC43520.1 GL888365 EGI62128.1 KQ982182 KYQ59161.1 KQ976514 KYM82219.1 ADTU01014356 KQ981410 KYN41895.1 PYGN01000327 PSN48446.1 KQ976750 KYN08615.1 KQ414627 KOC67416.1 UFQS01004045 UFQT01004045 SSX16125.1 SSX35452.1 GL447910 EFN85709.1 LBMM01004552 KMQ92298.1 DS235832 EEB18098.1 GL763739 EFZ19357.1 GL437123 EFN70998.1 AJWK01005820 KK107199 QOIP01000009 EZA55542.1 RLU18968.1 GECZ01005318 JAS64451.1 NNAY01000303 OXU29351.1 GECU01027521 GECU01000504 JAS80185.1 JAT07203.1 GEDC01012547 JAS24751.1 GBHO01016245 GDHC01009495 GDHC01008261 JAG27359.1 JAQ09134.1 JAQ10368.1 CCAG010014067 GBGD01002455 JAC86434.1 KQ435828 KOX71842.1
KPJ10250.1 GAIX01004063 JAA88497.1 ODYU01000407 SOQ35135.1 AGBW02014349 OWR41830.1 KQ459592 KPI96867.1 GANO01001402 JAB58469.1 DS232327 EDS40110.1 GFDL01010605 JAV24440.1 GBXI01002377 JAD11915.1 CH933816 EDW17278.1 CH940647 EDW68686.1 AAAB01008960 EAA11794.3 GDHF01005901 JAI46413.1 KA648111 AFP62740.1 GAMC01001923 JAC04633.1 GFDF01008417 JAV05667.1 APCN01004405 AXCM01002989 AXCN02000586 GAKP01004374 JAC54578.1 ATLV01023258 KE525341 KFB48738.1 ADMH02001932 ETN60443.1 JRES01000240 KNC33041.1 GAPW01002956 JAC10642.1 GGFM01004053 MBW24804.1 GGFK01009826 MBW43147.1 GGFJ01007911 MBW57052.1 JXUM01105523 KQ564973 KXJ71485.1 CH916366 EDV95871.1 JXUM01073321 KQ562767 KXJ75165.1 CH379070 EAL30097.1 CM000159 EDW93396.1 CH479203 EDW30346.1 APGK01010618 APGK01017343 BT127604 KB740012 KB738240 KB631654 AEE62566.1 ENN81901.1 ENN82710.1 ERL85053.1 CH902618 EDV38953.1 OUUW01000002 SPP77163.1 GDAI01000766 JAI16837.1 CH964278 EDW85354.1 CH477766 EAT36460.1 CH954178 EDV50907.1 U72893 AE014296 BT022197 AAD00188.1 AAF47880.1 AAY51591.1 CH480817 EDW50464.1 CM000363 CM002912 EDX09203.1 KMY97561.1 KQ971354 EFA05897.1 EZ422985 ADD19261.1 ODYU01001544 SOQ37910.1 JTDY01001830 KOB72772.1 GEZM01013039 JAV92801.1 KZ288223 PBC32023.1 JXJN01002265 NEVH01012562 PNF30239.1 KQ434795 KZC05528.1 CP012525 ALC43520.1 GL888365 EGI62128.1 KQ982182 KYQ59161.1 KQ976514 KYM82219.1 ADTU01014356 KQ981410 KYN41895.1 PYGN01000327 PSN48446.1 KQ976750 KYN08615.1 KQ414627 KOC67416.1 UFQS01004045 UFQT01004045 SSX16125.1 SSX35452.1 GL447910 EFN85709.1 LBMM01004552 KMQ92298.1 DS235832 EEB18098.1 GL763739 EFZ19357.1 GL437123 EFN70998.1 AJWK01005820 KK107199 QOIP01000009 EZA55542.1 RLU18968.1 GECZ01005318 JAS64451.1 NNAY01000303 OXU29351.1 GECU01027521 GECU01000504 JAS80185.1 JAT07203.1 GEDC01012547 JAS24751.1 GBHO01016245 GDHC01009495 GDHC01008261 JAG27359.1 JAQ09134.1 JAQ10368.1 CCAG010014067 GBGD01002455 JAC86434.1 KQ435828 KOX71842.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000002320
+ More
UP000009192 UP000008792 UP000075882 UP000076407 UP000007062 UP000075880 UP000095301 UP000075903 UP000075840 UP000075900 UP000075901 UP000075883 UP000075886 UP000075884 UP000030765 UP000076408 UP000000673 UP000037069 UP000075881 UP000075920 UP000069272 UP000069940 UP000249989 UP000095300 UP000192221 UP000001070 UP000001819 UP000002282 UP000075885 UP000008744 UP000019118 UP000030742 UP000007801 UP000268350 UP000007798 UP000008820 UP000008711 UP000000803 UP000001292 UP000000304 UP000007266 UP000037510 UP000242457 UP000092460 UP000235965 UP000092443 UP000076502 UP000092553 UP000007755 UP000075809 UP000078540 UP000005205 UP000078541 UP000245037 UP000078542 UP000053825 UP000008237 UP000036403 UP000009046 UP000000311 UP000005203 UP000092461 UP000053097 UP000279307 UP000215335 UP000002358 UP000092444 UP000075902 UP000053105 UP000092445
UP000009192 UP000008792 UP000075882 UP000076407 UP000007062 UP000075880 UP000095301 UP000075903 UP000075840 UP000075900 UP000075901 UP000075883 UP000075886 UP000075884 UP000030765 UP000076408 UP000000673 UP000037069 UP000075881 UP000075920 UP000069272 UP000069940 UP000249989 UP000095300 UP000192221 UP000001070 UP000001819 UP000002282 UP000075885 UP000008744 UP000019118 UP000030742 UP000007801 UP000268350 UP000007798 UP000008820 UP000008711 UP000000803 UP000001292 UP000000304 UP000007266 UP000037510 UP000242457 UP000092460 UP000235965 UP000092443 UP000076502 UP000092553 UP000007755 UP000075809 UP000078540 UP000005205 UP000078541 UP000245037 UP000078542 UP000053825 UP000008237 UP000036403 UP000009046 UP000000311 UP000005203 UP000092461 UP000053097 UP000279307 UP000215335 UP000002358 UP000092444 UP000075902 UP000053105 UP000092445
Pfam
Interpro
IPR016656
TFIIE-bsu
+ More
IPR040501 TFA2_Winged_2
IPR036388 WH-like_DNA-bd_sf
IPR036390 WH_DNA-bd_sf
IPR003166 TFIIE_bsu_DNA-bd
IPR000210 BTB/POZ_dom
IPR011333 SKP1/BTB/POZ_sf
IPR029019 HEX_eukaryotic_N
IPR015883 Glyco_hydro_20_cat
IPR029018 Hex-like_dom2
IPR017853 Glycoside_hydrolase_SF
IPR025705 Beta_hexosaminidase_sua/sub
IPR040501 TFA2_Winged_2
IPR036388 WH-like_DNA-bd_sf
IPR036390 WH_DNA-bd_sf
IPR003166 TFIIE_bsu_DNA-bd
IPR000210 BTB/POZ_dom
IPR011333 SKP1/BTB/POZ_sf
IPR029019 HEX_eukaryotic_N
IPR015883 Glyco_hydro_20_cat
IPR029018 Hex-like_dom2
IPR017853 Glycoside_hydrolase_SF
IPR025705 Beta_hexosaminidase_sua/sub
Gene 3D
ProteinModelPortal
H9J8N6
A0A2A4JI13
A0A1E1WHF1
A0A0N1IFX0
S4PY18
A0A2H1V2R2
+ More
A0A212EK30 A0A194PU71 U5EUM5 B0X433 A0A1Q3FA59 A0A0A1XM27 B4L9A3 B4LBQ9 A0A182KW59 A0A182XCZ9 Q7Q7C7 A0A182IKD0 A0A0K8W5D1 T1PH62 W8BZ93 A0A1L8DH07 A0A182UXT4 A0A182HMC7 A0A182R2N9 A0A182SB97 A0A182LZD4 A0A182QG40 A0A182N7T1 A0A034WLD0 A0A084WEU4 A0A182YNP2 W5JAL9 A0A0L0CLB8 A0A023EMR2 A0A182KAI9 A0A182WAN9 A0A2M3Z8F2 A0A2M4AQS9 A0A182FFQ2 A0A2M4BVN0 A0A182H2L8 A0A1I8Q164 A0A1W4VUN4 B4J1I0 A0A182GLZ8 Q29EG4 B4PHT7 A0A182PUU5 B4H1Z4 J3JWI6 B3M909 A0A3B0J9U2 A0A0K8TR33 B4NLY6 A0A1S4FTA4 Q16Q16 B3NC19 O96881 B4HU43 B4QQJ7 D6WSM2 D3TNI3 A0A2H1VAM8 A0A0L7LBA6 A0A1Y1N4G3 A0A2A3EJN2 A0A1B0AR63 A0A2J7QNS9 A0A1A9XGS6 A0A154P2S5 A0A0M4EYZ6 F4WUG3 A0A151XFP8 A0A195BDD9 A0A158NFE3 A0A151JZH5 A0A2P8YW15 A0A195D6U3 A0A0L7R9I5 A0A336LHH7 E2BF31 A0A0J7KPL3 E0VXJ2 E9IJ90 E2A6B3 A0A087ZQE1 A0A1B0CBT6 A0A026WHK3 A0A1B6GPV1 A0A232FF13 A0A1B6HZY4 K7JAG0 A0A1B6DGD1 A0A0A9Y5A9 A0A1B0FRJ7 A0A069DYD8 A0A182UI79 A0A0N0BEF6 A0A1A9ZG27
A0A212EK30 A0A194PU71 U5EUM5 B0X433 A0A1Q3FA59 A0A0A1XM27 B4L9A3 B4LBQ9 A0A182KW59 A0A182XCZ9 Q7Q7C7 A0A182IKD0 A0A0K8W5D1 T1PH62 W8BZ93 A0A1L8DH07 A0A182UXT4 A0A182HMC7 A0A182R2N9 A0A182SB97 A0A182LZD4 A0A182QG40 A0A182N7T1 A0A034WLD0 A0A084WEU4 A0A182YNP2 W5JAL9 A0A0L0CLB8 A0A023EMR2 A0A182KAI9 A0A182WAN9 A0A2M3Z8F2 A0A2M4AQS9 A0A182FFQ2 A0A2M4BVN0 A0A182H2L8 A0A1I8Q164 A0A1W4VUN4 B4J1I0 A0A182GLZ8 Q29EG4 B4PHT7 A0A182PUU5 B4H1Z4 J3JWI6 B3M909 A0A3B0J9U2 A0A0K8TR33 B4NLY6 A0A1S4FTA4 Q16Q16 B3NC19 O96881 B4HU43 B4QQJ7 D6WSM2 D3TNI3 A0A2H1VAM8 A0A0L7LBA6 A0A1Y1N4G3 A0A2A3EJN2 A0A1B0AR63 A0A2J7QNS9 A0A1A9XGS6 A0A154P2S5 A0A0M4EYZ6 F4WUG3 A0A151XFP8 A0A195BDD9 A0A158NFE3 A0A151JZH5 A0A2P8YW15 A0A195D6U3 A0A0L7R9I5 A0A336LHH7 E2BF31 A0A0J7KPL3 E0VXJ2 E9IJ90 E2A6B3 A0A087ZQE1 A0A1B0CBT6 A0A026WHK3 A0A1B6GPV1 A0A232FF13 A0A1B6HZY4 K7JAG0 A0A1B6DGD1 A0A0A9Y5A9 A0A1B0FRJ7 A0A069DYD8 A0A182UI79 A0A0N0BEF6 A0A1A9ZG27
PDB
6O9L
E-value=2.46158e-57,
Score=561
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
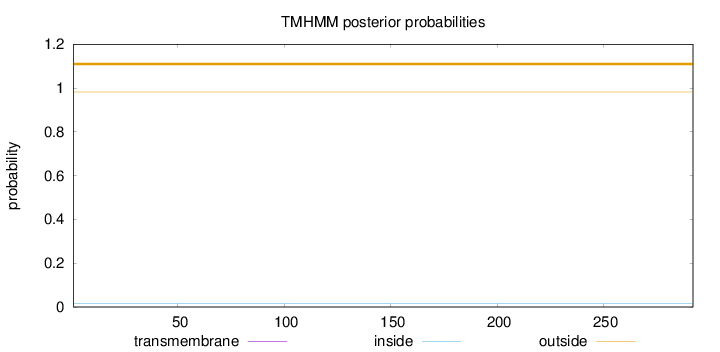
Length:
292
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00036
Exp number, first 60 AAs:
9e-05
Total prob of N-in:
0.01758
outside
1 - 292
Population Genetic Test Statistics
Pi
20.741146
Theta
19.949439
Tajima's D
0.423513
CLR
0.5725
CSRT
0.497025148742563
Interpretation
Uncertain
