Gene
KWMTBOMO07501 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005899
Annotation
PREDICTED:_chitooligosaccharidolytic_beta-N-acetylglucosaminidase_isoform_X1_[Bombyx_mori]
Full name
Chitooligosaccharidolytic beta-N-acetylglucosaminidase
+ More
Beta-hexosaminidase
Beta-hexosaminidase
Alternative Name
Beta-GlcNAcase
Beta-N-acetylhexosaminidase
Beta-hexosaminidase
Beta-N-acetylhexosaminidase
Beta-hexosaminidase
Location in the cell
Cytoplasmic Reliability : 1.389
Sequence
CDS
ATGAGAAAACTCAATGAGCAATCATATAAAAATGACAGATTCCAAATTCAAATTCCGACGGCAGCCGAAGAGCACTCATTGTGGAGATGGACGTGCGAAAACAATAGATGCACGAAGATCAGGAACGAGCCGGAGAATAAAGAGCCTGTCCTCAGTCTGGAAGCATGCAAGATGTTCTGTGACGATTACGGTCTCCTCTGGCCGAAACCAACGATCGAGACAAATCTGGGTAACTTCCTCTCCAAGATTAATATGAACACCATCGACATTCAGATCACCAAGCAGGGAAAGAGCGACGACCTCCTCACAGCGGCTGCCGACAGATTTAAGACACTGGTTTCCAGTTCCGTGCCAAAAGGCTTCTCGGCGAAGGCTGCAGGAAAATCAGTCACAGTTTATTTAGTCAATGACAATCCTTATATCCGAGAATTCTCCCTGGACATGGATGAGAGCTACGAGCTCTACATTTCATCAACGTCGAGCGACAAAGTGAACGCTACCATACGTGGAAACTCTTTCTTCGGCGTCCGTCACGGTCTGGAGACGCTCTCTCAGCTAATCGTGTACGACGACATAAGGAATAATCTGTTGATTGTCCGTGACGTCACAATTAAGGACAGGCCTGTCTATCCTTATCGAGGTATTTTGTTGGACACCGCGAGGAACTTCTACTCCATAGAATCAATCAAAAGAACTATAGACGCAATGGCCGCAGTGAAACTGAACACATTCCACTGGCACATCACGGACAGCCAGAGTTTCCCGTTGGTGTTACAGAAAAGACCGAACTTGAGTAAGCTCGGGGCATACAGTCCCACCAAGGTGTACACAAAGCAAGATATTCGTGAAGTTGTCGCATATGGTCTAGAGAGGGGAGTTCGTGTTTTACCGGAATTTGATGCGCCTGCTCATGTGGGAGAGGGATGGCAGGACACTGGACTCACCGTGTGCTTTAAGGCGGAGCCGTGGACGAAATTCTGCGTGGAGCCGCCTTGTGGTCAGCTGAACCCGACTAAAGAGGAACTTTACGACTATTTGGAAGATATTTACGTTGAAATGGCTGAGGCGTTTGAGAGCACCGACATGTTCCACATGGGAGGAGACGAGGTCAGCGAACGCTGTTGGAACTCCTCAGAGGAGATCCAGAACTTTATGATTCAAAACCGATGGAATTTGGACAAGAGCAGTTTCTTGAAGCTTTGGAACTACTTCCAGAAGAACGCTCAAGACAGAGCGTATAAGGCCTTCGGTAAACGACTGCCTCTGATTCTATGGACCAGCACATTGACCGACTACACTCACGTAGAGAAATTCTTGGACAAAGACGAATACATCATACAGGTCTGGACCACGGGAGCCGACCCACAAATTCAAGGTTTACTCCAGAAAGGATATCGCCTGATTATGTCAAATTATGACGCTCTGTACTTTGACTGTGGATTCGGGGCATGGGTTGGGTCCGGCAATAATTGGTGCTCACCGTACATCGGGTGGCAGAAAGTGTATGGTAACAGTCCAGCGGTGATGGCGCTCTCGTACCGAGATCAGATCTTGGGTGGTGAAGTAGCGCTGTGGTCCGAGCAGTCAGACCCTGCCACGCTGGACGGCCGACTGTGGCCCAGAGCGGCTGCCTTCGCCGAGCGCATGTGGGCCGAACCTTCCACCGCGTGGCAGGACGCCGAGCACCGGATGCTCCATGTCAGAGAACGTTTGGTAAGAATGGGAATTCAAGCTGAATCGCTTGAGCCGGAGTGGTGCTATCAGAATCAAGGACTTTGCTATGGTTAG
Protein
MRKLNEQSYKNDRFQIQIPTAAEEHSLWRWTCENNRCTKIRNEPENKEPVLSLEACKMFCDDYGLLWPKPTIETNLGNFLSKINMNTIDIQITKQGKSDDLLTAAADRFKTLVSSSVPKGFSAKAAGKSVTVYLVNDNPYIREFSLDMDESYELYISSTSSDKVNATIRGNSFFGVRHGLETLSQLIVYDDIRNNLLIVRDVTIKDRPVYPYRGILLDTARNFYSIESIKRTIDAMAAVKLNTFHWHITDSQSFPLVLQKRPNLSKLGAYSPTKVYTKQDIREVVAYGLERGVRVLPEFDAPAHVGEGWQDTGLTVCFKAEPWTKFCVEPPCGQLNPTKEELYDYLEDIYVEMAEAFESTDMFHMGGDEVSERCWNSSEEIQNFMIQNRWNLDKSSFLKLWNYFQKNAQDRAYKAFGKRLPLILWTSTLTDYTHVEKFLDKDEYIIQVWTTGADPQIQGLLQKGYRLIMSNYDALYFDCGFGAWVGSGNNWCSPYIGWQKVYGNSPAVMALSYRDQILGGEVALWSEQSDPATLDGRLWPRAAAFAERMWAEPSTAWQDAEHRMLHVRERLVRMGIQAESLEPEWCYQNQGLCYG
Summary
Catalytic Activity
Hydrolysis of terminal non-reducing N-acetyl-D-hexosamine residues in N-acetyl-beta-D-hexosaminides.
Similarity
Belongs to the glycosyl hydrolase 20 family.
Keywords
Carbohydrate metabolism
Chitin degradation
Complete proteome
Direct protein sequencing
Glycoprotein
Glycosidase
Hydrolase
Polysaccharide degradation
Reference proteome
Signal
Feature
chain Chitooligosaccharidolytic beta-N-acetylglucosaminidase
Uniprot
P49010
Q9GPG8
H9J8Q7
Q6UJX7
A0A2A4JK08
Q06GJ0
+ More
A0A0K0Q482 Q8T4N1 A0A218N0E0 D6PX67 Q52H16 A0A194PTX4 A0A0N1INV6 R4X5G1 C5IBZ5 A0A0F6PA61 A0A0L7LJ05 Q0MUV1 A0A3G1VUC7 A0A0T6BFY9 A0A0K8TL07 D6WPS5 A5YVX3 A0A1Y1MFL2 A0A3G5BIK2 A0A336MGX8 A0A182HB96 A0A1S4F898 Q17C82 A0A1J1IDP2 A0A1Y1MFQ5 A0A1Y1MF42 W8C231 T1PGB1 A0A182PUU4 A0A182WAN8 A0A182XD00 A0A0A1WSJ6 B4NLY5 A0A182M4F8 B4PHT8 A0A182KW66 A0A0L0CQ06 W8C2W8 A0A0K8U2M5 A0A182VK01 Q7Q7C8 A0A0R1E1S0 A0A182TF75 A0A0K8V7G3 A0A182HMC8 A0A084WEU5 B4LBR0 A0A0K8W4A2 A0A034VMN1 A0A182KAJ0 A0A0A1WY49 A0A0K8WHW6 A0A1W4VUL9 B4L9A2 A0A1W4W769 A0A2P0P9I9 A0A182SF49 A0A0K8VUH2 B4H1Z3 A0A0J9UEF6 Q0E8H9 Q8IRB6 A0A0J9RNW8 A0A182R2N8 B3M910 Q29EG3 B4HU44 B0X434 Q8MSS9 A0A067RD91 A0A2J7Q7A7 B4QQJ8 A0A182H8G3 A0A034VP89 A0A0A1WVT1 B4J1I1 A0A182N7T2 A0A182YNP1 K7J7Q3 A0A1W4XGX0 A0A3B0JAY0 A0A0K8VWT2 B3NC20 A0A0Q5UI80 A0A1L8E1Y0 A0A2M4DMQ0 A0A2M4DNW7 T1EAY4 A0A1L8E1N2 A0A182QJN0 A0A1A9WD41 A0A182FFQ0 A0A0M4ED89
A0A0K0Q482 Q8T4N1 A0A218N0E0 D6PX67 Q52H16 A0A194PTX4 A0A0N1INV6 R4X5G1 C5IBZ5 A0A0F6PA61 A0A0L7LJ05 Q0MUV1 A0A3G1VUC7 A0A0T6BFY9 A0A0K8TL07 D6WPS5 A5YVX3 A0A1Y1MFL2 A0A3G5BIK2 A0A336MGX8 A0A182HB96 A0A1S4F898 Q17C82 A0A1J1IDP2 A0A1Y1MFQ5 A0A1Y1MF42 W8C231 T1PGB1 A0A182PUU4 A0A182WAN8 A0A182XD00 A0A0A1WSJ6 B4NLY5 A0A182M4F8 B4PHT8 A0A182KW66 A0A0L0CQ06 W8C2W8 A0A0K8U2M5 A0A182VK01 Q7Q7C8 A0A0R1E1S0 A0A182TF75 A0A0K8V7G3 A0A182HMC8 A0A084WEU5 B4LBR0 A0A0K8W4A2 A0A034VMN1 A0A182KAJ0 A0A0A1WY49 A0A0K8WHW6 A0A1W4VUL9 B4L9A2 A0A1W4W769 A0A2P0P9I9 A0A182SF49 A0A0K8VUH2 B4H1Z3 A0A0J9UEF6 Q0E8H9 Q8IRB6 A0A0J9RNW8 A0A182R2N8 B3M910 Q29EG3 B4HU44 B0X434 Q8MSS9 A0A067RD91 A0A2J7Q7A7 B4QQJ8 A0A182H8G3 A0A034VP89 A0A0A1WVT1 B4J1I1 A0A182N7T2 A0A182YNP1 K7J7Q3 A0A1W4XGX0 A0A3B0JAY0 A0A0K8VWT2 B3NC20 A0A0Q5UI80 A0A1L8E1Y0 A0A2M4DMQ0 A0A2M4DNW7 T1EAY4 A0A1L8E1N2 A0A182QJN0 A0A1A9WD41 A0A182FFQ0 A0A0M4ED89
EC Number
3.2.1.52
Pubmed
7766021
19121390
8763162
18959754
21692744
21106526
+ More
23300622 25155420 30135205 18330895 26354079 26227816 26369729 18362917 19820115 18342252 28004739 30400621 26483478 17510324 24495485 25315136 25830018 17994087 17550304 20966253 26108605 12364791 14747013 17210077 24438588 25348373 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 15632085 24845553 25244985 20075255
23300622 25155420 30135205 18330895 26354079 26227816 26369729 18362917 19820115 18342252 28004739 30400621 26483478 17510324 24495485 25315136 25830018 17994087 17550304 20966253 26108605 12364791 14747013 17210077 24438588 25348373 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 15632085 24845553 25244985 20075255
EMBL
S77548
AF326597
AAG48701.1
BABH01030382
BABH01030383
BABH01030384
+ More
BABH01030385 BABH01030386 AY368703 AAQ97603.1 NWSH01001195 PCG72159.1 DQ887769 ABI81756.1 KP730442 AKR06190.1 AY078172 AAL82580.1 KY348777 ASF79655.1 GU985280 ADF56765.1 DQ005717 AAX94571.1 KQ459592 KPI96866.1 KQ460954 KPJ10249.1 AB766061 BAN29056.1 FJ848784 ACR57832.1 KJ632673 AJF93429.1 JTDY01000943 KOB75344.1 DQ845101 ABH09730.1 MH027208 AYK27452.1 LJIG01001141 KRT85799.1 GDAI01002541 JAI15062.1 KQ971354 EFA06863.2 EF592536 ABQ95982.1 GEZM01032380 GEZM01032379 JAV84619.1 MK075198 AYV99601.1 UFQT01001058 SSX28661.1 JXUM01124582 JXUM01124583 JXUM01124584 CH477311 EAT43909.1 CVRI01000047 CRK98399.1 GEZM01033085 JAV84421.1 GEZM01033084 JAV84422.1 GAMC01003317 JAC03239.1 KA646918 AFP61547.1 GBXI01012904 JAD01388.1 CH964278 EDW85353.2 AXCM01002989 CM000159 EDW93397.1 JRES01000080 KNC34346.1 GAMC01006349 JAC00207.1 GDHF01031593 JAI20721.1 AAAB01008960 EAA10994.4 KRK01159.1 GDHF01017447 JAI34867.1 APCN01004405 ATLV01023258 KE525341 KFB48739.1 CH940647 EDW68687.1 GDHF01006388 JAI45926.1 GAKP01014361 JAC44591.1 GBXI01010530 JAD03762.1 GDHF01001591 GDHF01001266 JAI50723.1 JAI51048.1 CH933816 EDW17277.2 KU739382 ARA90559.1 GDHF01010119 GDHF01007292 JAI42195.1 JAI45022.1 CH479203 EDW30345.1 CM002912 KMY97565.1 AE014296 AAF47881.1 BT099589 AAG22248.2 AAN11596.1 ACU43551.1 KMY97563.1 CH902618 EDV38954.2 KPU77762.1 CH379070 EAL30098.1 CH480817 EDW50465.1 DS232327 EDS40111.1 AY118626 AAM49995.1 KK852777 KDR16738.1 NEVH01017447 PNF24460.1 CM000363 EDX09204.1 JXUM01118347 JXUM01118348 GAKP01013831 JAC45121.1 GBXI01011118 JAD03174.1 CH916366 EDV95872.1 AAZX01006692 OUUW01000002 SPP77162.1 GDHF01009289 JAI43025.1 CH954178 EDV50908.1 KQS43574.1 GFDF01001445 JAV12639.1 GGFL01014631 MBW78809.1 GGFL01014630 MBW78808.1 GAMD01000643 JAB00948.1 GFDF01001444 JAV12640.1 AXCN02000586 CP012525 ALC43521.1
BABH01030385 BABH01030386 AY368703 AAQ97603.1 NWSH01001195 PCG72159.1 DQ887769 ABI81756.1 KP730442 AKR06190.1 AY078172 AAL82580.1 KY348777 ASF79655.1 GU985280 ADF56765.1 DQ005717 AAX94571.1 KQ459592 KPI96866.1 KQ460954 KPJ10249.1 AB766061 BAN29056.1 FJ848784 ACR57832.1 KJ632673 AJF93429.1 JTDY01000943 KOB75344.1 DQ845101 ABH09730.1 MH027208 AYK27452.1 LJIG01001141 KRT85799.1 GDAI01002541 JAI15062.1 KQ971354 EFA06863.2 EF592536 ABQ95982.1 GEZM01032380 GEZM01032379 JAV84619.1 MK075198 AYV99601.1 UFQT01001058 SSX28661.1 JXUM01124582 JXUM01124583 JXUM01124584 CH477311 EAT43909.1 CVRI01000047 CRK98399.1 GEZM01033085 JAV84421.1 GEZM01033084 JAV84422.1 GAMC01003317 JAC03239.1 KA646918 AFP61547.1 GBXI01012904 JAD01388.1 CH964278 EDW85353.2 AXCM01002989 CM000159 EDW93397.1 JRES01000080 KNC34346.1 GAMC01006349 JAC00207.1 GDHF01031593 JAI20721.1 AAAB01008960 EAA10994.4 KRK01159.1 GDHF01017447 JAI34867.1 APCN01004405 ATLV01023258 KE525341 KFB48739.1 CH940647 EDW68687.1 GDHF01006388 JAI45926.1 GAKP01014361 JAC44591.1 GBXI01010530 JAD03762.1 GDHF01001591 GDHF01001266 JAI50723.1 JAI51048.1 CH933816 EDW17277.2 KU739382 ARA90559.1 GDHF01010119 GDHF01007292 JAI42195.1 JAI45022.1 CH479203 EDW30345.1 CM002912 KMY97565.1 AE014296 AAF47881.1 BT099589 AAG22248.2 AAN11596.1 ACU43551.1 KMY97563.1 CH902618 EDV38954.2 KPU77762.1 CH379070 EAL30098.1 CH480817 EDW50465.1 DS232327 EDS40111.1 AY118626 AAM49995.1 KK852777 KDR16738.1 NEVH01017447 PNF24460.1 CM000363 EDX09204.1 JXUM01118347 JXUM01118348 GAKP01013831 JAC45121.1 GBXI01011118 JAD03174.1 CH916366 EDV95872.1 AAZX01006692 OUUW01000002 SPP77162.1 GDHF01009289 JAI43025.1 CH954178 EDV50908.1 KQS43574.1 GFDF01001445 JAV12639.1 GGFL01014631 MBW78809.1 GGFL01014630 MBW78808.1 GAMD01000643 JAB00948.1 GFDF01001444 JAV12640.1 AXCN02000586 CP012525 ALC43521.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000007266
+ More
UP000069940 UP000008820 UP000183832 UP000095301 UP000075885 UP000075920 UP000076407 UP000007798 UP000075883 UP000002282 UP000075882 UP000037069 UP000075903 UP000007062 UP000075902 UP000075840 UP000030765 UP000008792 UP000075881 UP000192221 UP000009192 UP000075901 UP000008744 UP000000803 UP000075900 UP000007801 UP000001819 UP000001292 UP000002320 UP000027135 UP000235965 UP000000304 UP000001070 UP000075884 UP000076408 UP000002358 UP000192223 UP000268350 UP000008711 UP000075886 UP000091820 UP000069272 UP000092553
UP000069940 UP000008820 UP000183832 UP000095301 UP000075885 UP000075920 UP000076407 UP000007798 UP000075883 UP000002282 UP000075882 UP000037069 UP000075903 UP000007062 UP000075902 UP000075840 UP000030765 UP000008792 UP000075881 UP000192221 UP000009192 UP000075901 UP000008744 UP000000803 UP000075900 UP000007801 UP000001819 UP000001292 UP000002320 UP000027135 UP000235965 UP000000304 UP000001070 UP000075884 UP000076408 UP000002358 UP000192223 UP000268350 UP000008711 UP000075886 UP000091820 UP000069272 UP000092553
Interpro
IPR029018
Hex-like_dom2
+ More
IPR017853 Glycoside_hydrolase_SF
IPR025705 Beta_hexosaminidase_sua/sub
IPR029019 HEX_eukaryotic_N
IPR015883 Glyco_hydro_20_cat
IPR027483 PInositol-4-P-5-kinase_C
IPR002498 PInositol-4-P-5-kinase_core
IPR036390 WH_DNA-bd_sf
IPR036388 WH-like_DNA-bd_sf
IPR003166 TFIIE_bsu_DNA-bd
IPR027484 PInositol-4-P-5-kinase_N
IPR017853 Glycoside_hydrolase_SF
IPR025705 Beta_hexosaminidase_sua/sub
IPR029019 HEX_eukaryotic_N
IPR015883 Glyco_hydro_20_cat
IPR027483 PInositol-4-P-5-kinase_C
IPR002498 PInositol-4-P-5-kinase_core
IPR036390 WH_DNA-bd_sf
IPR036388 WH-like_DNA-bd_sf
IPR003166 TFIIE_bsu_DNA-bd
IPR027484 PInositol-4-P-5-kinase_N
Gene 3D
ProteinModelPortal
P49010
Q9GPG8
H9J8Q7
Q6UJX7
A0A2A4JK08
Q06GJ0
+ More
A0A0K0Q482 Q8T4N1 A0A218N0E0 D6PX67 Q52H16 A0A194PTX4 A0A0N1INV6 R4X5G1 C5IBZ5 A0A0F6PA61 A0A0L7LJ05 Q0MUV1 A0A3G1VUC7 A0A0T6BFY9 A0A0K8TL07 D6WPS5 A5YVX3 A0A1Y1MFL2 A0A3G5BIK2 A0A336MGX8 A0A182HB96 A0A1S4F898 Q17C82 A0A1J1IDP2 A0A1Y1MFQ5 A0A1Y1MF42 W8C231 T1PGB1 A0A182PUU4 A0A182WAN8 A0A182XD00 A0A0A1WSJ6 B4NLY5 A0A182M4F8 B4PHT8 A0A182KW66 A0A0L0CQ06 W8C2W8 A0A0K8U2M5 A0A182VK01 Q7Q7C8 A0A0R1E1S0 A0A182TF75 A0A0K8V7G3 A0A182HMC8 A0A084WEU5 B4LBR0 A0A0K8W4A2 A0A034VMN1 A0A182KAJ0 A0A0A1WY49 A0A0K8WHW6 A0A1W4VUL9 B4L9A2 A0A1W4W769 A0A2P0P9I9 A0A182SF49 A0A0K8VUH2 B4H1Z3 A0A0J9UEF6 Q0E8H9 Q8IRB6 A0A0J9RNW8 A0A182R2N8 B3M910 Q29EG3 B4HU44 B0X434 Q8MSS9 A0A067RD91 A0A2J7Q7A7 B4QQJ8 A0A182H8G3 A0A034VP89 A0A0A1WVT1 B4J1I1 A0A182N7T2 A0A182YNP1 K7J7Q3 A0A1W4XGX0 A0A3B0JAY0 A0A0K8VWT2 B3NC20 A0A0Q5UI80 A0A1L8E1Y0 A0A2M4DMQ0 A0A2M4DNW7 T1EAY4 A0A1L8E1N2 A0A182QJN0 A0A1A9WD41 A0A182FFQ0 A0A0M4ED89
A0A0K0Q482 Q8T4N1 A0A218N0E0 D6PX67 Q52H16 A0A194PTX4 A0A0N1INV6 R4X5G1 C5IBZ5 A0A0F6PA61 A0A0L7LJ05 Q0MUV1 A0A3G1VUC7 A0A0T6BFY9 A0A0K8TL07 D6WPS5 A5YVX3 A0A1Y1MFL2 A0A3G5BIK2 A0A336MGX8 A0A182HB96 A0A1S4F898 Q17C82 A0A1J1IDP2 A0A1Y1MFQ5 A0A1Y1MF42 W8C231 T1PGB1 A0A182PUU4 A0A182WAN8 A0A182XD00 A0A0A1WSJ6 B4NLY5 A0A182M4F8 B4PHT8 A0A182KW66 A0A0L0CQ06 W8C2W8 A0A0K8U2M5 A0A182VK01 Q7Q7C8 A0A0R1E1S0 A0A182TF75 A0A0K8V7G3 A0A182HMC8 A0A084WEU5 B4LBR0 A0A0K8W4A2 A0A034VMN1 A0A182KAJ0 A0A0A1WY49 A0A0K8WHW6 A0A1W4VUL9 B4L9A2 A0A1W4W769 A0A2P0P9I9 A0A182SF49 A0A0K8VUH2 B4H1Z3 A0A0J9UEF6 Q0E8H9 Q8IRB6 A0A0J9RNW8 A0A182R2N8 B3M910 Q29EG3 B4HU44 B0X434 Q8MSS9 A0A067RD91 A0A2J7Q7A7 B4QQJ8 A0A182H8G3 A0A034VP89 A0A0A1WVT1 B4J1I1 A0A182N7T2 A0A182YNP1 K7J7Q3 A0A1W4XGX0 A0A3B0JAY0 A0A0K8VWT2 B3NC20 A0A0Q5UI80 A0A1L8E1Y0 A0A2M4DMQ0 A0A2M4DNW7 T1EAY4 A0A1L8E1N2 A0A182QJN0 A0A1A9WD41 A0A182FFQ0 A0A0M4ED89
PDB
3S6T
E-value=0,
Score=2489
Ontologies
PATHWAY
00511
Other glycan degradation - Bombyx mori (domestic silkworm)
00520 Amino sugar and nucleotide sugar metabolism - Bombyx mori (domestic silkworm)
00531 Glycosaminoglycan degradation - Bombyx mori (domestic silkworm)
00603 Glycosphingolipid biosynthesis - globo and isoglobo series - Bombyx mori (domestic silkworm)
00604 Glycosphingolipid biosynthesis - ganglio series - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
04142 Lysosome - Bombyx mori (domestic silkworm)
00520 Amino sugar and nucleotide sugar metabolism - Bombyx mori (domestic silkworm)
00531 Glycosaminoglycan degradation - Bombyx mori (domestic silkworm)
00603 Glycosphingolipid biosynthesis - globo and isoglobo series - Bombyx mori (domestic silkworm)
00604 Glycosphingolipid biosynthesis - ganglio series - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
04142 Lysosome - Bombyx mori (domestic silkworm)
GO
Topology
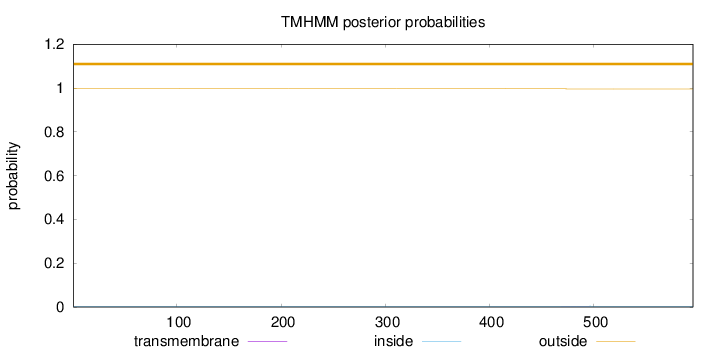
Length:
595
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01601
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00342
outside
1 - 595
Population Genetic Test Statistics
Pi
21.39565
Theta
21.589699
Tajima's D
-0.929023
CLR
0.862514
CSRT
0.151342432878356
Interpretation
Uncertain
