Gene
KWMTBOMO07498 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005876
Annotation
PREDICTED:_low_density_lipoprotein_receptor_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.688
Sequence
CDS
ATGTGGACGTGCGCTGGTTTGCTCGCGTTGGCCACGTGCGCCGTCGCGATTACCCGGGACCAGTTCTACCCTTATGGGCCGGAGTTGGACCAGAGGTTGCCCAGGGGATCGGAAGTCCCATCGCCGGAAGTCACTCTGAACGTCCCTATCGTATTCTACGATGAAACCTACGAGACAGTTTTTGTGAACAACCACGGCGTGCTAACGTTCCGCGCAAATCTCCCTCAATTTATCAACACGGAATTCCCACTGCCATACCCAGCAATAGCGGCGTTCTACTCCAACGTCGACACCACCGACGAAGGAGCAATCTACTACAGAGAGACCAACGAATCTCACATCCTATCAAAGGCTGAAGAAAGCGTGCAGAACAACTTTCACGATTACTATGACTTCCGACCAACGAGTGTCTTCATAACAACTTGGCTGGACGTCACTTACACAAGCACTTTACACGACAGAAAGAACAGTTACCAAGTCGCTGTGATCAGTAATGGAACGGAAAGTTTTGTTGAGCTACTATATCCGGAGAGAGAGATCCAGTGGATTCAGAAAGAGACGGATGCACTGTCGCTGCCGGACGCTAAAGCCCAAGCTGGTTTCGTGGCGGAGGACGGCCGCGTCTTCACCCTTCGTGGCTCGGGCAGTCATCAAATCAGAAATATTATATCATGGACAAACACACACGAGCCTGGAAAATACGTCTTCCGTGTTGGCACTATACCTCGCGACTCAAACATCGCTGTTCCTGATGAATACAATGAGACCGAAATTGAAATTGCGGAAGAGTCCAAAACGTGCGCCCAGAGCGGTCCCAGCGTATGCCACATGCAAGCGAGATGCGTAGACTACCAGGCCGGCATCTGCTGTCAATGCAGCGAAGGATTCTACGGGAACGGAAAATCATGCATTAAAAACGACGTACCTCTGAGAGTACACGGAAAACTGAATGGAGTTCTGAATGACGTGCACCTAAATGATGTGGATATCCAGTCTTACGTTGTAGTTGCTGACGGGAGAGCTTACACGGCACTGTCTCTTACTCCACCAGTCCTCGGAAGCAGTTTGCAGTTGCTAAACATCATGGGCAGCGTAGTTGGCTGGCTGTTTGCGAAGCCATCTTCCACGGCCAAAAACGGATACCAATTAACAGGCTCGCTTTTCAACCACAGCGTTGACATTTGGTTCCCGAAAACCGGTGACAGGGTTACCGTAAACCAAGAATACTTAGGTCACGATGTTTTCGACCAAATAACTTTAGACGCTGATGTACGTGGCTCTGTGCCGGTTATTTTATCCGGAATCAAACTAGACATTGGAGACTTTGAAGAGCAATATACCATTGTCGAACCGGGACTTATTCGTTCCGATTCTAGTCGAACGTTCACAAATAAGATCACAGGAGAAACGTACGAACAAAGAGTGTCCCAGACCATCACCTATAGGGCGTGTCGATACGCGCCTCCCTCAGCCGATGACACAATTCCATCCACTCTTAAAGTCACCAAAAACTATTTAGGTTATGAAACGAAGAACAATATAGTAAGATACGGAACGAGCAACAAGATTGTACCGCTAGGCCAAGAAGATCCCTGTATCGAAGGTCGTAACACTTGCGGTTCGTACAGCACGTGTGTCGTCCAAGGTGATTCCTTTAGCTGCGTTTGTAATAAAGGATTTAACAAGATTTATGTTGACGATGTGCTGACCTGTGTCGACATTGATGAATGCGCAGCCGGAACACATAACTGCGATCCAAACGCTGACTGCAACAACGGGGAAGGAGACTTTCAATGTCGATGCAAAACTGGATTCATTGGCAACGGAATCGAATGTTCACCGTTGTCTATATCTCGATGCCAAAGTCTGAGTTGCGATGTGAACGCGGACTGTTTAGAGAACAATGGAGAAGAACCGATATGCGTGTGCAGACCCGGTTACACTGGTAACGGGCAACAGTGCTATGAAGTCGATGAGCGCGGATGTTACGAAGGCTCCGGCTGCACTGAAAGACCTCCAGAGACAACCACGGCGCCTAACTACGAATCGGAATATAACGAAACATTCGTGTTGCCTAACTGCGACTATTACGGCTGCACGTGCCCCCAAGGCTACTACAGTTTCAAAGATGAGAGAAACAACGATCTGTGTCGTGTTGATACAGGCGACGTGCCGACCCCTATCACCGATAACAATGACACTTCAATCAGGTGCGATAGCGACAGTGACTGCCCACCAAACGCTGCTTGTATAGCGGACCCACATCAATACTCAACGGATGCTGAGAGCGATCAGTTCCTCGGACACTGCGTTTGCCCTGAAGGCTACGTGGGCGATGGATACGAATGCATCGAGCAAACCGGAACCAGCTGCAATTGTGCAGCCAACGCTCATTGTACGGAAACGCCAACTGCGGAACTTCTATGCGTCTGCAATACCGGTTACCACGGAGACGGCTACGTCTGCCGTCCGAATTTCAGTTGTACAAACAACTCGGACTGTGAATACAACGCTGAATGCCGTCCCGATACTGCCACAAACGAATACACTTGTCAGTGCATCGAAGGATTTAAAAAGGACCAAAATGAAGCTTGCATCCCTGACGGTCAATTGTGTAATGGCGCGCTGTGTGCTGAGCACGCCTCGTGCTTGTACGATGCCACCATTGAAATCAGTTACTGCCATTGCGACGAGGGCTTTAACGGCGAAGGCATATCCCAATGCGTCCAGACCCCACTCTCGTGTGACCTAGTCAACGACTGCAGTCCGTACGCGAGCTGCGTGCCAGCTGATAATACTTACCAGTGCGTGTGCCAAGAGGGCTATCTGGGCGACGGCTACAACTGCAGTCAGGAAATCAACTGCAGAACTGATCCTTACATGTGCAGTCCTCACGCTTCGTGCATAAAGAACATTGAAGGTTACGAATGTGAATGCAATAATGCATACGAGGGTAACGGCACTTTCTGCGTTGCGAAACCTCAGCCGCCGGGTAACTTTTTAGTGACAAGTGATGGAACTTCAATCTACCGCGTCCCGTTCAAGTATTCACCGAGGGAGTTTGCAACTCCGATTAACAGCGGCATTTCCCAAGTGGCAGTTTCCGTGAGCGTCGATTGTTTCTCTGGAAAAATCTACTGGGGCGATGTTGTCAGCAATAACATCAAACGAGCTGGATACGACGGATCAGAATTTGAACAATTCCTGAACACTGGCATCATATCTCCGGAAGGTTTGGCAATAGACTGGTCGGCTAGAAACATATTTTGGACAGATTCTAAGAAGCTCACTATTGAAGTTGCCAATTTAGACACAAAAGTTAGAAAAGTCTTGTTCTCGGTACAGAACATTTCTAATCCGAGGGGCATCGCCGTACATCCTCAAAGAGGGAAAATCTTCTGGACTGACTGGAATCGAAGCAATCCTAAGATCGAGTGGGCGAACATGGACGGCACGGAAAGATCAGTCCTCCTCGGTAGCCCAGATGTGAAGCTTCCAAACTCTGTGGCCATCGACTGGTCGAGAGATCGAGTTTGCTTCGCGGACGCGGGCCTCCAGGTCATTAAGTGCATTGACCTACACAGTCGGCAAATCGAGACTATTGCGGAGAACTGCAGGTACCCCTTCGGCCTCGCCATCAACGGAGACAAATTGTACTGGAGCGATTGGAGAACATTGAAAATCGAAATCATAGACACAATAACCCAGGCGAGAGGGCAAATAGACATTCCAAGATCGACGATACGGTTATACGGTGTTGCCAACGCGCCGAGCGAGTGTCCCGGGGCTGCCAACGTTTGCTCGCAGAGAAACGGTAACTGCAACGCCGGACAGCTGTGCCTACCCAATGGAAGTGGGGGAAGGAGTTGTCTGAACGGTGACGCCAACTATTTACCTTAG
Protein
MWTCAGLLALATCAVAITRDQFYPYGPELDQRLPRGSEVPSPEVTLNVPIVFYDETYETVFVNNHGVLTFRANLPQFINTEFPLPYPAIAAFYSNVDTTDEGAIYYRETNESHILSKAEESVQNNFHDYYDFRPTSVFITTWLDVTYTSTLHDRKNSYQVAVISNGTESFVELLYPEREIQWIQKETDALSLPDAKAQAGFVAEDGRVFTLRGSGSHQIRNIISWTNTHEPGKYVFRVGTIPRDSNIAVPDEYNETEIEIAEESKTCAQSGPSVCHMQARCVDYQAGICCQCSEGFYGNGKSCIKNDVPLRVHGKLNGVLNDVHLNDVDIQSYVVVADGRAYTALSLTPPVLGSSLQLLNIMGSVVGWLFAKPSSTAKNGYQLTGSLFNHSVDIWFPKTGDRVTVNQEYLGHDVFDQITLDADVRGSVPVILSGIKLDIGDFEEQYTIVEPGLIRSDSSRTFTNKITGETYEQRVSQTITYRACRYAPPSADDTIPSTLKVTKNYLGYETKNNIVRYGTSNKIVPLGQEDPCIEGRNTCGSYSTCVVQGDSFSCVCNKGFNKIYVDDVLTCVDIDECAAGTHNCDPNADCNNGEGDFQCRCKTGFIGNGIECSPLSISRCQSLSCDVNADCLENNGEEPICVCRPGYTGNGQQCYEVDERGCYEGSGCTERPPETTTAPNYESEYNETFVLPNCDYYGCTCPQGYYSFKDERNNDLCRVDTGDVPTPITDNNDTSIRCDSDSDCPPNAACIADPHQYSTDAESDQFLGHCVCPEGYVGDGYECIEQTGTSCNCAANAHCTETPTAELLCVCNTGYHGDGYVCRPNFSCTNNSDCEYNAECRPDTATNEYTCQCIEGFKKDQNEACIPDGQLCNGALCAEHASCLYDATIEISYCHCDEGFNGEGISQCVQTPLSCDLVNDCSPYASCVPADNTYQCVCQEGYLGDGYNCSQEINCRTDPYMCSPHASCIKNIEGYECECNNAYEGNGTFCVAKPQPPGNFLVTSDGTSIYRVPFKYSPREFATPINSGISQVAVSVSVDCFSGKIYWGDVVSNNIKRAGYDGSEFEQFLNTGIISPEGLAIDWSARNIFWTDSKKLTIEVANLDTKVRKVLFSVQNISNPRGIAVHPQRGKIFWTDWNRSNPKIEWANMDGTERSVLLGSPDVKLPNSVAIDWSRDRVCFADAGLQVIKCIDLHSRQIETIAENCRYPFGLAINGDKLYWSDWRTLKIEIIDTITQARGQIDIPRSTIRLYGVANAPSECPGAANVCSQRNGNCNAGQLCLPNGSGGRSCLNGDANYLP
Summary
Uniprot
H9J8N4
A0A1E1WIG0
A0A212FM33
A0A3S2NFD0
A0A2H1VBC9
A0A0L7LIT9
+ More
D1ZZG7 A0A2J7R912 A0A067QW96 A0A1Y1LZY0 A0A026X154 A0A1Y1M364 A0A0T6AUY8 A0A3L8DPT4 A0A2A3EIC9 A0A087ZZ55 A0A158NLJ2 A0A195B211 E2BGJ0 A0A310SQF9 A0A232EVQ4 A0A0L7QTG1 A0A151IB75 A0A0C9R1C5 A0A0N0BFG6 A0A154PD98 A0A151WG69 U4UHW1 F4WW20 E2B0R2 T1JGD6 E9G6S4 A0A195EXZ6 A0A1B6D9X9 A0A0P5UWB8 B0WBL6 A0A1B6BY70 A0A0P5DC43 A0A0P5TBY2 A0A0P4YNS9 A0A0P5YG05 A0A0P6GTF9 A0A0P5U4F4 A0A1Q3FIC5 A0A0P5YFI9 A0A0P5T7D5 A0A0P5CV86 A0A182YME0 A0A0P5UXN2 A0A0P5CMH1 A0A0N8A2M8 A0A0P5B768 A0A0P5WW64 A0A182RCN5 A0A0P4Z907 A0A182M8K2 A0A0N7ZPM2 A0A182QJE3 A0A182NFC0 A0A0P6IA20 A0A182XKY2 A0A182L179 A0A182TNS0 A0A182VE75 A0A182HUH2 A0A182JJM2 A0A1S4GXK2 A0A0P5QAA7 A0A0P6CH78 A0A182KA77 A0A0P5YYE7 A0A182W6W5 W5JF32 A0A182PPQ1 A0A0P5VRM5 A0A0P5VB08 A0A0P5W503 A0A182F202 Q7Q440 A0A084VL71 A0A182GF26 A0A1S4FG03 Q172X3 A0A1B0GHS0 A0A1J1HNG2 A0A1B6MHX2 W8B8M3 T1I1L9 A0A1B6MH41 A0A0A9X8D5 A0A146LSX4 A0A1B0DDW6 A0A132ACX8 A0A336M830 A0A0A1WSH1 A0A034V1L5 R7TE17
D1ZZG7 A0A2J7R912 A0A067QW96 A0A1Y1LZY0 A0A026X154 A0A1Y1M364 A0A0T6AUY8 A0A3L8DPT4 A0A2A3EIC9 A0A087ZZ55 A0A158NLJ2 A0A195B211 E2BGJ0 A0A310SQF9 A0A232EVQ4 A0A0L7QTG1 A0A151IB75 A0A0C9R1C5 A0A0N0BFG6 A0A154PD98 A0A151WG69 U4UHW1 F4WW20 E2B0R2 T1JGD6 E9G6S4 A0A195EXZ6 A0A1B6D9X9 A0A0P5UWB8 B0WBL6 A0A1B6BY70 A0A0P5DC43 A0A0P5TBY2 A0A0P4YNS9 A0A0P5YG05 A0A0P6GTF9 A0A0P5U4F4 A0A1Q3FIC5 A0A0P5YFI9 A0A0P5T7D5 A0A0P5CV86 A0A182YME0 A0A0P5UXN2 A0A0P5CMH1 A0A0N8A2M8 A0A0P5B768 A0A0P5WW64 A0A182RCN5 A0A0P4Z907 A0A182M8K2 A0A0N7ZPM2 A0A182QJE3 A0A182NFC0 A0A0P6IA20 A0A182XKY2 A0A182L179 A0A182TNS0 A0A182VE75 A0A182HUH2 A0A182JJM2 A0A1S4GXK2 A0A0P5QAA7 A0A0P6CH78 A0A182KA77 A0A0P5YYE7 A0A182W6W5 W5JF32 A0A182PPQ1 A0A0P5VRM5 A0A0P5VB08 A0A0P5W503 A0A182F202 Q7Q440 A0A084VL71 A0A182GF26 A0A1S4FG03 Q172X3 A0A1B0GHS0 A0A1J1HNG2 A0A1B6MHX2 W8B8M3 T1I1L9 A0A1B6MH41 A0A0A9X8D5 A0A146LSX4 A0A1B0DDW6 A0A132ACX8 A0A336M830 A0A0A1WSH1 A0A034V1L5 R7TE17
Pubmed
EMBL
BABH01030389
BABH01030390
GDQN01004251
JAT86803.1
AGBW02007661
OWR54787.1
+ More
RSAL01000060 RVE49650.1 ODYU01001622 SOQ38106.1 JTDY01000943 KOB75340.1 KQ971338 EFA02368.1 NEVH01006721 PNF37315.1 KK852870 KDR14602.1 GEZM01043436 JAV79119.1 KK107063 EZA61094.1 GEZM01043435 JAV79120.1 LJIG01022752 KRT78905.1 QOIP01000005 RLU22252.1 KZ288231 PBC31553.1 ADTU01019501 ADTU01019502 ADTU01019503 ADTU01019504 KQ976662 KYM78327.1 GL448189 EFN85183.1 KQ761010 OAD58397.1 NNAY01001967 OXU22427.1 KQ414743 KOC61928.1 KQ978125 KYM96871.1 GBYB01006682 JAG76449.1 KQ435804 KOX73048.1 KQ434878 KZC09875.1 KQ983190 KYQ46808.1 KB632308 ERL91928.1 GL888404 EGI61609.1 GL444666 EFN60734.1 JH432201 GL732533 EFX84793.1 KQ981914 KYN33098.1 GEDC01014795 JAS22503.1 GDIP01107377 JAL96337.1 DS231880 EDS42551.1 GEDC01031066 JAS06232.1 GDIP01159814 JAJ63588.1 GDIP01130171 JAL73543.1 GDIP01224962 GDIP01206013 GDIP01047084 LRGB01000337 JAI98439.1 KZS19650.1 GDIP01058435 JAM45280.1 GDIQ01039199 JAN55538.1 GDIP01118074 JAL85640.1 GFDL01007747 JAV27298.1 GDIP01058434 JAM45281.1 GDIP01136476 JAL67238.1 GDIP01169211 JAJ54191.1 GDIP01107376 JAL96338.1 GDIP01168365 JAJ55037.1 GDIP01188513 JAJ34889.1 GDIP01188514 JAJ34888.1 GDIP01081128 JAM22587.1 GDIP01217712 JAJ05690.1 AXCM01002282 GDIP01224961 JAI98440.1 AXCN02000820 GDIQ01007467 JAN87270.1 APCN01002461 AAAB01008964 GDIQ01117130 JAL34596.1 GDIP01008721 JAM94994.1 GDIP01064795 JAM38920.1 ADMH02001591 ETN61938.1 GDIP01096381 JAM07334.1 GDIP01104723 JAL98991.1 GDIP01104722 JAL98992.1 EAA12454.4 ATLV01014417 KE524970 KFB38715.1 JXUM01010366 JXUM01010367 KQ560305 KXJ83140.1 CH477429 EAT41106.1 AJWK01005550 AJWK01005551 AJWK01005552 AJWK01005553 AJWK01005554 AJWK01005555 CVRI01000013 CRK89591.1 GEBQ01004486 JAT35491.1 GAMC01020427 GAMC01020426 JAB86129.1 ACPB03011058 GEBQ01004707 JAT35270.1 GBHO01027703 JAG15901.1 GDHC01020959 GDHC01010289 GDHC01007751 JAP97669.1 JAQ08340.1 JAQ10878.1 AJVK01005477 AJVK01005478 JXLN01012783 KPM08846.1 UFQT01000527 SSX25003.1 GBXI01012701 JAD01591.1 GAKP01021806 JAC37146.1 AMQN01013652 KB310369 ELT91752.1
RSAL01000060 RVE49650.1 ODYU01001622 SOQ38106.1 JTDY01000943 KOB75340.1 KQ971338 EFA02368.1 NEVH01006721 PNF37315.1 KK852870 KDR14602.1 GEZM01043436 JAV79119.1 KK107063 EZA61094.1 GEZM01043435 JAV79120.1 LJIG01022752 KRT78905.1 QOIP01000005 RLU22252.1 KZ288231 PBC31553.1 ADTU01019501 ADTU01019502 ADTU01019503 ADTU01019504 KQ976662 KYM78327.1 GL448189 EFN85183.1 KQ761010 OAD58397.1 NNAY01001967 OXU22427.1 KQ414743 KOC61928.1 KQ978125 KYM96871.1 GBYB01006682 JAG76449.1 KQ435804 KOX73048.1 KQ434878 KZC09875.1 KQ983190 KYQ46808.1 KB632308 ERL91928.1 GL888404 EGI61609.1 GL444666 EFN60734.1 JH432201 GL732533 EFX84793.1 KQ981914 KYN33098.1 GEDC01014795 JAS22503.1 GDIP01107377 JAL96337.1 DS231880 EDS42551.1 GEDC01031066 JAS06232.1 GDIP01159814 JAJ63588.1 GDIP01130171 JAL73543.1 GDIP01224962 GDIP01206013 GDIP01047084 LRGB01000337 JAI98439.1 KZS19650.1 GDIP01058435 JAM45280.1 GDIQ01039199 JAN55538.1 GDIP01118074 JAL85640.1 GFDL01007747 JAV27298.1 GDIP01058434 JAM45281.1 GDIP01136476 JAL67238.1 GDIP01169211 JAJ54191.1 GDIP01107376 JAL96338.1 GDIP01168365 JAJ55037.1 GDIP01188513 JAJ34889.1 GDIP01188514 JAJ34888.1 GDIP01081128 JAM22587.1 GDIP01217712 JAJ05690.1 AXCM01002282 GDIP01224961 JAI98440.1 AXCN02000820 GDIQ01007467 JAN87270.1 APCN01002461 AAAB01008964 GDIQ01117130 JAL34596.1 GDIP01008721 JAM94994.1 GDIP01064795 JAM38920.1 ADMH02001591 ETN61938.1 GDIP01096381 JAM07334.1 GDIP01104723 JAL98991.1 GDIP01104722 JAL98992.1 EAA12454.4 ATLV01014417 KE524970 KFB38715.1 JXUM01010366 JXUM01010367 KQ560305 KXJ83140.1 CH477429 EAT41106.1 AJWK01005550 AJWK01005551 AJWK01005552 AJWK01005553 AJWK01005554 AJWK01005555 CVRI01000013 CRK89591.1 GEBQ01004486 JAT35491.1 GAMC01020427 GAMC01020426 JAB86129.1 ACPB03011058 GEBQ01004707 JAT35270.1 GBHO01027703 JAG15901.1 GDHC01020959 GDHC01010289 GDHC01007751 JAP97669.1 JAQ08340.1 JAQ10878.1 AJVK01005477 AJVK01005478 JXLN01012783 KPM08846.1 UFQT01000527 SSX25003.1 GBXI01012701 JAD01591.1 GAKP01021806 JAC37146.1 AMQN01013652 KB310369 ELT91752.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000037510
UP000007266
UP000235965
+ More
UP000027135 UP000053097 UP000279307 UP000242457 UP000005203 UP000005205 UP000078540 UP000008237 UP000215335 UP000053825 UP000078542 UP000053105 UP000076502 UP000075809 UP000030742 UP000007755 UP000000311 UP000000305 UP000078541 UP000002320 UP000076858 UP000076408 UP000075900 UP000075883 UP000075886 UP000075884 UP000076407 UP000075882 UP000075902 UP000075903 UP000075840 UP000075880 UP000075881 UP000075920 UP000000673 UP000075885 UP000069272 UP000007062 UP000030765 UP000069940 UP000249989 UP000008820 UP000092461 UP000183832 UP000015103 UP000092462 UP000014760
UP000027135 UP000053097 UP000279307 UP000242457 UP000005203 UP000005205 UP000078540 UP000008237 UP000215335 UP000053825 UP000078542 UP000053105 UP000076502 UP000075809 UP000030742 UP000007755 UP000000311 UP000000305 UP000078541 UP000002320 UP000076858 UP000076408 UP000075900 UP000075883 UP000075886 UP000075884 UP000076407 UP000075882 UP000075902 UP000075903 UP000075840 UP000075880 UP000075881 UP000075920 UP000000673 UP000075885 UP000069272 UP000007062 UP000030765 UP000069940 UP000249989 UP000008820 UP000092461 UP000183832 UP000015103 UP000092462 UP000014760
PRIDE
Pfam
Interpro
IPR000742
EGF-like_dom
+ More
IPR011042 6-blade_b-propeller_TolB-like
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR003886 NIDO_dom
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR024731 EGF_dom
IPR000033 LDLR_classB_rpt
IPR001881 EGF-like_Ca-bd_dom
IPR009030 Growth_fac_rcpt_cys_sf
IPR006605 G2_nidogen/fibulin_G2F
IPR009017 GFP
IPR011992 EF-hand-dom_pair
IPR000195 Rab-GTPase-TBC_dom
IPR021935 SGSM1/2_RBD
IPR035969 Rab-GTPase_TBC_sf
IPR034754 GEMIN8
IPR024730 MSP1_EGF_1
IPR011042 6-blade_b-propeller_TolB-like
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR003886 NIDO_dom
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR024731 EGF_dom
IPR000033 LDLR_classB_rpt
IPR001881 EGF-like_Ca-bd_dom
IPR009030 Growth_fac_rcpt_cys_sf
IPR006605 G2_nidogen/fibulin_G2F
IPR009017 GFP
IPR011992 EF-hand-dom_pair
IPR000195 Rab-GTPase-TBC_dom
IPR021935 SGSM1/2_RBD
IPR035969 Rab-GTPase_TBC_sf
IPR034754 GEMIN8
IPR024730 MSP1_EGF_1
Gene 3D
CDD
ProteinModelPortal
H9J8N4
A0A1E1WIG0
A0A212FM33
A0A3S2NFD0
A0A2H1VBC9
A0A0L7LIT9
+ More
D1ZZG7 A0A2J7R912 A0A067QW96 A0A1Y1LZY0 A0A026X154 A0A1Y1M364 A0A0T6AUY8 A0A3L8DPT4 A0A2A3EIC9 A0A087ZZ55 A0A158NLJ2 A0A195B211 E2BGJ0 A0A310SQF9 A0A232EVQ4 A0A0L7QTG1 A0A151IB75 A0A0C9R1C5 A0A0N0BFG6 A0A154PD98 A0A151WG69 U4UHW1 F4WW20 E2B0R2 T1JGD6 E9G6S4 A0A195EXZ6 A0A1B6D9X9 A0A0P5UWB8 B0WBL6 A0A1B6BY70 A0A0P5DC43 A0A0P5TBY2 A0A0P4YNS9 A0A0P5YG05 A0A0P6GTF9 A0A0P5U4F4 A0A1Q3FIC5 A0A0P5YFI9 A0A0P5T7D5 A0A0P5CV86 A0A182YME0 A0A0P5UXN2 A0A0P5CMH1 A0A0N8A2M8 A0A0P5B768 A0A0P5WW64 A0A182RCN5 A0A0P4Z907 A0A182M8K2 A0A0N7ZPM2 A0A182QJE3 A0A182NFC0 A0A0P6IA20 A0A182XKY2 A0A182L179 A0A182TNS0 A0A182VE75 A0A182HUH2 A0A182JJM2 A0A1S4GXK2 A0A0P5QAA7 A0A0P6CH78 A0A182KA77 A0A0P5YYE7 A0A182W6W5 W5JF32 A0A182PPQ1 A0A0P5VRM5 A0A0P5VB08 A0A0P5W503 A0A182F202 Q7Q440 A0A084VL71 A0A182GF26 A0A1S4FG03 Q172X3 A0A1B0GHS0 A0A1J1HNG2 A0A1B6MHX2 W8B8M3 T1I1L9 A0A1B6MH41 A0A0A9X8D5 A0A146LSX4 A0A1B0DDW6 A0A132ACX8 A0A336M830 A0A0A1WSH1 A0A034V1L5 R7TE17
D1ZZG7 A0A2J7R912 A0A067QW96 A0A1Y1LZY0 A0A026X154 A0A1Y1M364 A0A0T6AUY8 A0A3L8DPT4 A0A2A3EIC9 A0A087ZZ55 A0A158NLJ2 A0A195B211 E2BGJ0 A0A310SQF9 A0A232EVQ4 A0A0L7QTG1 A0A151IB75 A0A0C9R1C5 A0A0N0BFG6 A0A154PD98 A0A151WG69 U4UHW1 F4WW20 E2B0R2 T1JGD6 E9G6S4 A0A195EXZ6 A0A1B6D9X9 A0A0P5UWB8 B0WBL6 A0A1B6BY70 A0A0P5DC43 A0A0P5TBY2 A0A0P4YNS9 A0A0P5YG05 A0A0P6GTF9 A0A0P5U4F4 A0A1Q3FIC5 A0A0P5YFI9 A0A0P5T7D5 A0A0P5CV86 A0A182YME0 A0A0P5UXN2 A0A0P5CMH1 A0A0N8A2M8 A0A0P5B768 A0A0P5WW64 A0A182RCN5 A0A0P4Z907 A0A182M8K2 A0A0N7ZPM2 A0A182QJE3 A0A182NFC0 A0A0P6IA20 A0A182XKY2 A0A182L179 A0A182TNS0 A0A182VE75 A0A182HUH2 A0A182JJM2 A0A1S4GXK2 A0A0P5QAA7 A0A0P6CH78 A0A182KA77 A0A0P5YYE7 A0A182W6W5 W5JF32 A0A182PPQ1 A0A0P5VRM5 A0A0P5VB08 A0A0P5W503 A0A182F202 Q7Q440 A0A084VL71 A0A182GF26 A0A1S4FG03 Q172X3 A0A1B0GHS0 A0A1J1HNG2 A0A1B6MHX2 W8B8M3 T1I1L9 A0A1B6MH41 A0A0A9X8D5 A0A146LSX4 A0A1B0DDW6 A0A132ACX8 A0A336M830 A0A0A1WSH1 A0A034V1L5 R7TE17
PDB
1NPE
E-value=5.177e-44,
Score=453
Ontologies
GO
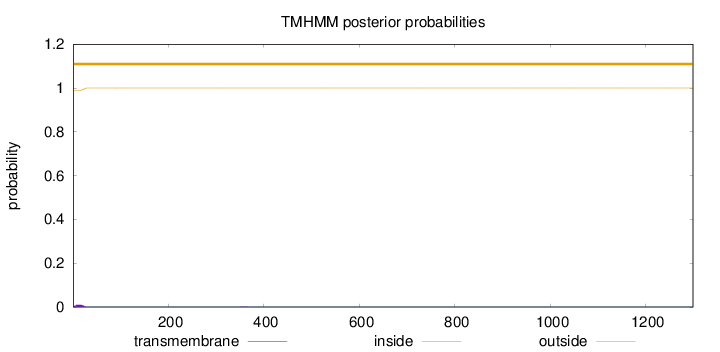
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.942121

Length:
1299
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.18933
Exp number, first 60 AAs:
0.18542
Total prob of N-in:
0.00988
outside
1 - 1299
Population Genetic Test Statistics
Pi
3.402475
Theta
14.680084
Tajima's D
-1.33936
CLR
18.469653
CSRT
0.079746012699365
Interpretation
Uncertain
