Gene
KWMTBOMO07497
Pre Gene Modal
BGIBMGA005875
Annotation
PREDICTED:_kinesin-like_protein_CG14535_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.457
Sequence
CDS
ATGCTGCGGGTTGGAGCGTCAGGAGATGGTCCTTTCACGAGCGGCAACCAGACCTTCGCCATGGACAAGCGGAAGAAACAAGTCAGCCTATGCGAAGCAACGTCGGCCGCTTCGGCTCCTGAAGACAGAAAAGTCGGAGTGACAGCTCCTAAGATGTTCGCCTTCGACGCTATTTTCTCGCAAGACGATCCACAGACGGAGATATGCTCGAGCGCCCTGACGGACGTGATTCACGCAGTCATCAACGGCACCGACGGATGCCTCTTTTGTTTCGGACACGCGGGGCTTGGTAAATCCTACACGATGCTGGGTCGGCCGGACTCGACACCGAATCTCGGCGCGATACCGTGCGCGATCACATGGCTGTTCCGAGGCATCGCAGAGCAGAGGCACAAGTGCGGCACAAGATTTTCCGTGCGAGTTTCGTGCGTGGAACTCTGCACAAATACAAATCAAATACGAGATCTGTTGGCGCCATATCAAAATGATACGGAGCAATCTCCAGGAGTCTACCTTCGAGACGATCCTCTTTTCGGTACCCACCTCCAGAACCAGTCAGAGCTTCGAGTTCACAGCGCAGAAAAGGCTGCATTTTATCTGGATGCGGCTCTAGCTACAAGGGGGATTAAAGAAGAGGGAAAAGATAGCCATTTGCTGTACACTCTACACGTTTACCAATACAGCGTTGGAGGGAAAGGAGCAGTGGCTGGGGGTCGAAGCCGTCTTCACCTCATTGACCTTGGAAACTCTGAACGTGGAAAAACGAACGGCGGCATTCCCCTTTCGGGATTAGGAAACATTCTGCTTGCTATTTTTAACGGTCAACGACATCTCCCTTATCGGGACCACAATCTAACTCACGTACTAAAAGAATGTTTAGGTTCTCTGACATGTCATACTGCCATGATAGTCCACGTGTCTCCTAACGTCCAAAACTACTCCGATACATTAACTACTCTACAATTAGCTTCAAGAATACACAGATTAAGGCGTAGAAAGCTCAAATACTCTTCGAATAACAATGCTGGTTCTGGTGGAAGCTCAGGCGAAGACGCTTCAAAACCGAGCAGCAGCGAACCTGATCCATCGAGTAGTGATTTATCAGCTGACACAGTAATATACGTGGGTCCTATGGACGATGCTACCGATGGTGAACATCCTCCTGTTTATATTCCTCATATTAACTCTGGAGACAACAGATGTATCCTTAGCAAAGCTTTAAGAGGCTCTGCAGCAGAGCAAAAGCATAAGTCTTCATTATCGAAGGTGGTTGAAGAGAAAAGTCCCATTCATAAATTACACTCGTCTCCTAAAGCATCTCCTTTGAAAACTATACAGCCCGGTCATACACCGAAACCATCTCCCGTTCATTCAGTTGCTTCAAAATCGACTCCTTTAAAAAAAGCTGCTCCAAAACAATACGTAGAAGACACTAAAAGTACTAGCGATGAACAGTGGATAGACGGGCCAAGGATATCGAAATCAAAATTGGTTGAGGCGAGACATATTATCAAGGAGACCCAAAATAAGAAACGTGAAACTTGGATTGATGGACCGATGCAAGAACCCAAACTCCCGCCACCATTTGACCCCTTAGCAATTCAACCCAATACGATACCCCTTCAAGTTGGAGTGCTAGGAGTTCACGGCAGTGAAAACATGGGTTATGGATATATGGATAGCCACAAAAAAAATATGATTCGAAAATGGGTCGAAAATCAAACATCACAAATCCACAAGTCCAGAAATAGCTCGCCAAGTCACAAGAATCATAAAGATAGCATTAAGACCGGAGATGAATCTGAAACTAACAGAATGGAAATAAAAGAAAGTGTCAGACGAGCTCATGTCGAAGAATCTACGATTAGAACCGGCTTGAAGGCTGGATCAAAAGGAAATATGATTGAAGAAGAAAATTTAGTATCATCGGAACAAAATGTAGTAGTGAAAAAATCTAACAGTAATAGTTCCAAAACAGGAAGAAATGATGTTGACGAAGACGATGATAGTGAAGAATTAGCAGAAATACCCCCAGCTTTGCCATTGATTCAGCCCAGCAGTCTGAATAGTCGAGAAGTGTCCATGGAGAGCTTAAATAGTAAATACAAGGAAAGATTAAGAATGGACGACGAAATGTTGTCTAATCACAGTAGAGAAATTGAGATTCACGGCATGAGTAGAGGCCATGAAGATGATGATGAAGATGAATTTTTAGAGATCATAGAAGTAGAAGAGCCTCTGGAGCCGGTTCCGATGCAGGACTGCTGCCTCCAAGTCACTGAGGAAGATATCGCTTTGTGTATGGGATTTGATAACACTTTACCCGGCGACCAAGACGATGATGATCACCCTCTCAGGATACTCAGTCAAGAGAATCTAACAGTTGCTTCAACATTTACGGACACTTTTTCCTTATACAGCGAAGTAGAAAGACAGATCAGAAAAGAAGCATATTTACGGCAAACCATGGAATTTTATACTGAAAACTTTACAAGCGATCAGCACCTCGGCGATCTGAATCCACATGAAAGTTTACATTCGTCGCGATCTAGAATAGACGACTACATAAACGATATTTACGAATCTCGACGAATGATAGAACACGATGAAATGATAAATACTAGAATCGCACCTCAATTCCAATCCATATCACTGTGTAATGTACGTGATACTGATGATTATCACGGCGATGGTTCTGTATATAGTGAACCAGCATATCGTCCTAGTGATAAGATTTGTGATAGTTGCAAACGAACCATGTCCAGGCCAGGGAGTGCTGTAGGTGGGGCTTATCACGAACCAGATGGATCTCACAACGACTGCTACGGGCCATTCGAAAGGTACAGAGGCAACGACATAACTGATGCAAGAATCGCATCTCTAAGACACCCAGATGGTGCATCCGACCCTAACTTGCGAGAAGAAAAACGAATACCCGGAAACGGGGCGTGTACTTTTAAAGAACTAAAATCTAATTTGCATTTGGAGGTGTCCCCTCCAGTTGTGACAACCGAACCTCGATCCGAGAAACATGAAAAGGGCGATAAATGCGTAGCCAGAATAGTTGCCGGATCGAAACCGGACGGATACGATAGCGGGCATGAATCAACTCCGAGAACCGGAAAACACAGTCCGGCCGCCACCTCGAGACGAGCCGAATCCGGCTACGACTCTGTTCCAAGAGACTCAGACGCGTCATCCTTGGACTCATACCCGACTAGACGAGCCGCAGTCGCACGGGCGCACGCTAAAAAACACACCACAGCGAAATACAAACAGCACAACGATCGTTCATTCTGCTCGTGGCTAAGGAATCCGTTCACGTGCAAATACGCTGACACCGATCCAGAAATCAGCGATTTTTAA
Protein
MLRVGASGDGPFTSGNQTFAMDKRKKQVSLCEATSAASAPEDRKVGVTAPKMFAFDAIFSQDDPQTEICSSALTDVIHAVINGTDGCLFCFGHAGLGKSYTMLGRPDSTPNLGAIPCAITWLFRGIAEQRHKCGTRFSVRVSCVELCTNTNQIRDLLAPYQNDTEQSPGVYLRDDPLFGTHLQNQSELRVHSAEKAAFYLDAALATRGIKEEGKDSHLLYTLHVYQYSVGGKGAVAGGRSRLHLIDLGNSERGKTNGGIPLSGLGNILLAIFNGQRHLPYRDHNLTHVLKECLGSLTCHTAMIVHVSPNVQNYSDTLTTLQLASRIHRLRRRKLKYSSNNNAGSGGSSGEDASKPSSSEPDPSSSDLSADTVIYVGPMDDATDGEHPPVYIPHINSGDNRCILSKALRGSAAEQKHKSSLSKVVEEKSPIHKLHSSPKASPLKTIQPGHTPKPSPVHSVASKSTPLKKAAPKQYVEDTKSTSDEQWIDGPRISKSKLVEARHIIKETQNKKRETWIDGPMQEPKLPPPFDPLAIQPNTIPLQVGVLGVHGSENMGYGYMDSHKKNMIRKWVENQTSQIHKSRNSSPSHKNHKDSIKTGDESETNRMEIKESVRRAHVEESTIRTGLKAGSKGNMIEEENLVSSEQNVVVKKSNSNSSKTGRNDVDEDDDSEELAEIPPALPLIQPSSLNSREVSMESLNSKYKERLRMDDEMLSNHSREIEIHGMSRGHEDDDEDEFLEIIEVEEPLEPVPMQDCCLQVTEEDIALCMGFDNTLPGDQDDDDHPLRILSQENLTVASTFTDTFSLYSEVERQIRKEAYLRQTMEFYTENFTSDQHLGDLNPHESLHSSRSRIDDYINDIYESRRMIEHDEMINTRIAPQFQSISLCNVRDTDDYHGDGSVYSEPAYRPSDKICDSCKRTMSRPGSAVGGAYHEPDGSHNDCYGPFERYRGNDITDARIASLRHPDGASDPNLREEKRIPGNGACTFKELKSNLHLEVSPPVVTTEPRSEKHEKGDKCVARIVAGSKPDGYDSGHESTPRTGKHSPAATSRRAESGYDSVPRDSDASSLDSYPTRRAAVARAHAKKHTTAKYKQHNDRSFCSWLRNPFTCKYADTDPEISDF
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Uniprot
Pubmed
EMBL
BABH01030397
BABH01030398
BABH01030399
BABH01030400
NWSH01002063
PCG69267.1
+ More
RSAL01000060 RVE49651.1 KQ460325 KPJ15742.1 KQ459592 KPI96863.1 AGBW02011193 OWR47074.1 ODYU01003439 SOQ42221.1 JTDY01000549 KOB76715.1 GEZM01038159 JAV81757.1 KQ971338 KYB27927.1 GL448189 EFN85182.1 KK107063 EZA61095.1 KQ414743 KOC61929.1 PYGN01001450 PSN34765.1 AAZX01003633 KQ761010 OAD58396.1 GEZM01038160 JAV81756.1 QOIP01000005 RLU22249.1 KK852449 KDR23709.1 APGK01031207 APGK01031208 KB740800 ENN78897.1 APGK01020423 APGK01020424 KB740198 ENN81055.1 OUUW01000004 SPP79167.1 CH479180 EDW29008.1
RSAL01000060 RVE49651.1 KQ460325 KPJ15742.1 KQ459592 KPI96863.1 AGBW02011193 OWR47074.1 ODYU01003439 SOQ42221.1 JTDY01000549 KOB76715.1 GEZM01038159 JAV81757.1 KQ971338 KYB27927.1 GL448189 EFN85182.1 KK107063 EZA61095.1 KQ414743 KOC61929.1 PYGN01001450 PSN34765.1 AAZX01003633 KQ761010 OAD58396.1 GEZM01038160 JAV81756.1 QOIP01000005 RLU22249.1 KK852449 KDR23709.1 APGK01031207 APGK01031208 KB740800 ENN78897.1 APGK01020423 APGK01020424 KB740198 ENN81055.1 OUUW01000004 SPP79167.1 CH479180 EDW29008.1
Proteomes
PRIDE
Pfam
PF00225 Kinesin
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
PDB
3B6U
E-value=1.69299e-26,
Score=301
Ontologies
GO
PANTHER
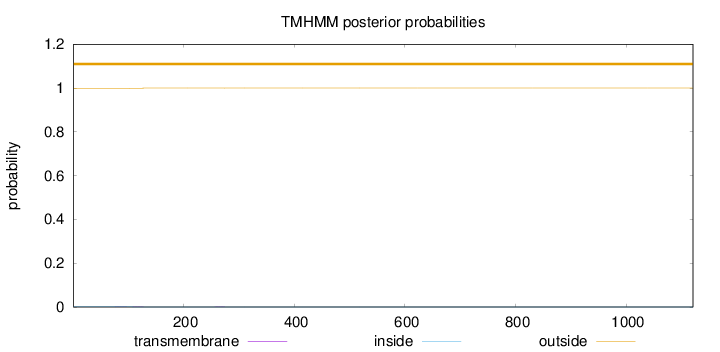
Topology
Length:
1121
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02492
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00107
outside
1 - 1121
Population Genetic Test Statistics
Pi
24.042357
Theta
23.428886
Tajima's D
-0.992861
CLR
1.263329
CSRT
0.133793310334483
Interpretation
Uncertain
