Gene
KWMTBOMO07494
Pre Gene Modal
BGIBMGA014556
Annotation
PREDICTED:_polypeptide_N-acetylgalactosaminyltransferase_2-like_isoform_X2_[Papilio_machaon]
Full name
Polypeptide N-acetylgalactosaminyltransferase
+ More
Polypeptide N-acetylgalactosaminyltransferase 2
Polypeptide N-acetylgalactosaminyltransferase 2
Alternative Name
Protein-UDP acetylgalactosaminyltransferase
Protein-UDP acetylgalactosaminyltransferase 2
UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 2
Protein-UDP acetylgalactosaminyltransferase 2
UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 2
Location in the cell
Cytoplasmic Reliability : 1.691 Mitochondrial Reliability : 1.469
Sequence
CDS
ATGGACACCTTCCAATACATCGGAGCGTCGGCGGACCTGCGAGGCGGTTTTGACTGGAACTTAGTCTTCAAGTGGGAATACTTATCGCATACGGAACGTAGCGCCCGTCTCACTGATCCCACTCAAGTAATCAGGACCCCTATGATAGCTGGTGGACTATTCAGCATGGACCGACTTTACTTCAACAAACTCGGGAAATACGATATGAAAATGGACGTGTGGGGAGGAGAGAACTTGGAAATTTCATTCAGAGTTTGGCAATGCGGAGGCTCCCTCGAAATCGTGCCTTGTTCCCGTGTGGGTCATGTCTTCAGGAAACGCCATCCGTATACATTCCCCGGCGGCTCCGGGGCGGTATTCGCGAGGAATACCAGAAGGGCGGCCGAAGTCTGGATGGACGATTACAAAGAACTATACTATAGATCTCAACCACTCGCGAAGCAAGTCGACTTTGGAGACATATCCGAGCGCGTCACTTTACGCGACAAACTACACTGCAAGCCGTTTCGTTGGTACTTACAACACGTGTACCCTGAACTCAAAATACCAGCCCAATTAGGGGGACCCCATCCTATAAAACAAGGAATAAGATGCCTAGACACCATGGGACATCTGGTCGACGGGACAATAGGAATGTATCCTTGCCATTACACGGGAGGCAATCAGGGGTGGCTCTTCGAAAACGGCCTCATCCGGCATCACAGTCTCTGCGTCTCCTTGTCCAACGAGGATCGTGTCACGGCAGTATTAGCACCCTGCGACCCTTCGGACGACGCCCAGCTCTGGACACGGAAGGGTCAACTCATAAGACACGAGAAACTGGACGCGTGCTTAGACACGGAACGCACTGATCTACATCTAGAACAATGCGACGAAGACAAACCGAGTCAGCAATTCACCTTTTAA
Protein
MDTFQYIGASADLRGGFDWNLVFKWEYLSHTERSARLTDPTQVIRTPMIAGGLFSMDRLYFNKLGKYDMKMDVWGGENLEISFRVWQCGGSLEIVPCSRVGHVFRKRHPYTFPGGSGAVFARNTRRAAEVWMDDYKELYYRSQPLAKQVDFGDISERVTLRDKLHCKPFRWYLQHVYPELKIPAQLGGPHPIKQGIRCLDTMGHLVDGTIGMYPCHYTGGNQGWLFENGLIRHHSLCVSLSNEDRVTAVLAPCDPSDDAQLWTRKGQLIRHEKLDACLDTERTDLHLEQCDEDKPSQQFTF
Summary
Catalytic Activity
L-seryl-[protein] + UDP-N-acetyl-alpha-D-galactosamine = 3-O-[N-acetyl-alpha-D-galactosaminyl]-L-seryl-[protein] + H(+) + UDP
L-threonyl-[protein] + UDP-N-acetyl-alpha-D-galactosamine = 3-O-[N-acetyl-alpha-D-galactosaminyl]-L-threonyl-[protein] + H(+) + UDP
L-threonyl-[protein] + UDP-N-acetyl-alpha-D-galactosamine = 3-O-[N-acetyl-alpha-D-galactosaminyl]-L-threonyl-[protein] + H(+) + UDP
Cofactor
Mn(2+)
Similarity
Belongs to the glycosyltransferase 2 family. GalNAc-T subfamily.
Keywords
Complete proteome
Disulfide bond
Glycosyltransferase
Golgi apparatus
Lectin
Manganese
Membrane
Metal-binding
Reference proteome
Signal-anchor
Transferase
Transmembrane
Transmembrane helix
Feature
chain Polypeptide N-acetylgalactosaminyltransferase 2
Uniprot
A0A2A4JVU1
A0A2H1V7K7
A0A194PVS1
H9JYD6
A0A194REM4
I4DLJ5
+ More
D2A0Q4 A0A232FKY8 T1I445 A0A195B151 F4WY07 A0A2A3EDA0 A0A088ANE0 A0A026WNX2 E2B4S0 A0A0N0BC47 A0A151J8G2 A0A023F4X4 A0A158NZ99 B4MWS9 A0A0M4E5A5 B3MNW1 A0A154PQG7 A0A1B6HAJ5 A0A1Y1LIC7 A0A151X933 B4KG89 A0A1B6L3A7 A0A195FV45 A0A3L8DW00 B4JDG8 A0A0K8VNP3 A0A3B0JZV2 A0A3B0K2J7 A0A1B6KJZ6 B4NXW1 B4G9P3 B3N316 Q29M65 A0A1W4UTS1 B4I2Z3 B4Q9J8 Q6WV19 A0A195CIK7 A0A1W4X6X2 B4LTQ9 A0A0L7RH75 A0A0J7KPE7 A0A0J9QV41 E2A7V1 A0A1S4EP92 A0A336LNH0 A0A0K8SYF1 A0A067R1I4 A0A0C9RCP2 A0A2R7W4N3 A0A0A9Y4T6 W8AN19 E9J4V7 A0A1B6C795 A0A1I8NIG4 A0A034V9M4 U4UH93 A0A1B0G2M0 A0A1B0A0V0 A0A1A9VTU0 A0A1A9Y369 A0A1B0BTJ8 A0A0L0BX46 A0A0K8V659 A0A0K8UCA0 A0A1I8NZS1 A0A1B6ISM0 A0A2P8Y6L9 A0A0K8WBU9 A0A0N7ZCW8 A0A1A9W0R7 Q16R35 E0W2D6 A0A182H3A2 B0WPN5 A0A0L7L238 A0A182V2F4 Q7Q0X9 A0A182XH81 A0A182PTI8 A0A182HR03 A0A182SZN9 A0A182TV63 A0A182Q6H4 A0A182JVQ3 A0A1Q3FPR8 A0A182LSH7 A0A1Q3FPW6 A0A210QER2 A0A182YQN2 A0A182NNR3 A0A182R1P7 W5JJQ0 A0A182W8H1
D2A0Q4 A0A232FKY8 T1I445 A0A195B151 F4WY07 A0A2A3EDA0 A0A088ANE0 A0A026WNX2 E2B4S0 A0A0N0BC47 A0A151J8G2 A0A023F4X4 A0A158NZ99 B4MWS9 A0A0M4E5A5 B3MNW1 A0A154PQG7 A0A1B6HAJ5 A0A1Y1LIC7 A0A151X933 B4KG89 A0A1B6L3A7 A0A195FV45 A0A3L8DW00 B4JDG8 A0A0K8VNP3 A0A3B0JZV2 A0A3B0K2J7 A0A1B6KJZ6 B4NXW1 B4G9P3 B3N316 Q29M65 A0A1W4UTS1 B4I2Z3 B4Q9J8 Q6WV19 A0A195CIK7 A0A1W4X6X2 B4LTQ9 A0A0L7RH75 A0A0J7KPE7 A0A0J9QV41 E2A7V1 A0A1S4EP92 A0A336LNH0 A0A0K8SYF1 A0A067R1I4 A0A0C9RCP2 A0A2R7W4N3 A0A0A9Y4T6 W8AN19 E9J4V7 A0A1B6C795 A0A1I8NIG4 A0A034V9M4 U4UH93 A0A1B0G2M0 A0A1B0A0V0 A0A1A9VTU0 A0A1A9Y369 A0A1B0BTJ8 A0A0L0BX46 A0A0K8V659 A0A0K8UCA0 A0A1I8NZS1 A0A1B6ISM0 A0A2P8Y6L9 A0A0K8WBU9 A0A0N7ZCW8 A0A1A9W0R7 Q16R35 E0W2D6 A0A182H3A2 B0WPN5 A0A0L7L238 A0A182V2F4 Q7Q0X9 A0A182XH81 A0A182PTI8 A0A182HR03 A0A182SZN9 A0A182TV63 A0A182Q6H4 A0A182JVQ3 A0A1Q3FPR8 A0A182LSH7 A0A1Q3FPW6 A0A210QER2 A0A182YQN2 A0A182NNR3 A0A182R1P7 W5JJQ0 A0A182W8H1
EC Number
2.4.1.-
2.4.1.41
2.4.1.41
Pubmed
26354079
19121390
22651552
18362917
19820115
28648823
+ More
21719571 24508170 20798317 25474469 21347285 17994087 18057021 28004739 30249741 17550304 15632085 23185243 22936249 10731132 12537572 12829714 16251381 24845553 25401762 26823975 24495485 21282665 25315136 25348373 23537049 26108605 29403074 17510324 20566863 26483478 26227816 12364791 28812685 25244985 20920257 23761445
21719571 24508170 20798317 25474469 21347285 17994087 18057021 28004739 30249741 17550304 15632085 23185243 22936249 10731132 12537572 12829714 16251381 24845553 25401762 26823975 24495485 21282665 25315136 25348373 23537049 26108605 29403074 17510324 20566863 26483478 26227816 12364791 28812685 25244985 20920257 23761445
EMBL
NWSH01000544
PCG75754.1
ODYU01001071
SOQ36797.1
KQ459592
KPI96859.1
+ More
BABH01043185 KQ460325 KPJ15740.1 AK402163 BAM18785.1 KQ971338 EFA01652.1 NNAY01000079 OXU31180.1 ACPB03003277 KQ976681 KYM78186.1 GL888434 EGI60888.1 KZ288276 PBC29725.1 KK107151 EZA57376.1 GL445595 EFN89299.1 KQ435944 KOX68172.1 KQ979528 KYN21147.1 GBBI01002658 JAC16054.1 ADTU01004134 ADTU01004135 ADTU01004136 ADTU01004137 ADTU01004138 ADTU01004139 ADTU01004140 ADTU01004141 ADTU01004142 ADTU01004143 CH963857 EDW76568.1 CP012523 ALC39246.1 CH902620 EDV32148.1 KPU73816.1 KQ435037 KZC14143.1 GECU01036013 JAS71693.1 GEZM01057944 JAV72080.1 KQ982392 KYQ56881.1 CH933807 EDW11076.1 KRG02521.1 GEBQ01021963 JAT18014.1 KQ981276 KYN43739.1 QOIP01000003 RLU24413.1 CH916368 EDW03338.1 GDHF01011810 JAI40504.1 OUUW01000004 SPP79229.1 SPP79231.1 GEBQ01028211 JAT11766.1 CM000157 EDW87532.1 CH479180 EDW29073.1 CH954177 EDV57615.1 CH379060 EAL33829.2 KRT04224.1 CH480820 EDW54138.1 CM000361 CM002910 EDX03609.1 KMY87884.1 AE014134 BT010030 AY268064 KQ977696 KYN00571.1 CH940649 EDW63960.1 KQ414596 KOC70061.1 LBMM01004751 KMQ92109.1 KMY87886.1 GL437408 EFN70488.1 UFQT01000093 SSX19622.1 GBRD01007886 GBRD01001372 JAG57935.1 KK852777 KDR16775.1 GBYB01004676 JAG74443.1 KK854326 PTY14687.1 GBHO01019049 GDHC01016936 JAG24555.1 JAQ01693.1 GAMC01020672 JAB85883.1 GL768119 EFZ12159.1 GEDC01027945 GEDC01011111 JAS09353.1 JAS26187.1 GAKP01020467 JAC38485.1 KB632299 ERL91738.1 CCAG010020175 JXJN01020204 JRES01001205 KNC24622.1 GDHF01017963 JAI34351.1 GDHF01028060 JAI24254.1 GECU01017809 JAS89897.1 PYGN01000865 PSN39900.1 GDHF01003678 JAI48636.1 GDRN01058336 JAI65608.1 CH477726 EAT36839.1 DS235877 EEB19792.1 JXUM01107019 JXUM01107020 JXUM01107021 JXUM01107022 JXUM01107023 JXUM01107024 JXUM01107025 KQ565104 KXJ71333.1 DS232027 EDS32438.1 JTDY01003479 KOB69495.1 AAAB01008980 EAA14535.4 APCN01001629 AXCN02001979 GFDL01005446 JAV29599.1 AXCM01000301 GFDL01005438 JAV29607.1 NEDP02003995 OWF47223.1 ADMH02001289 ETN63135.1
BABH01043185 KQ460325 KPJ15740.1 AK402163 BAM18785.1 KQ971338 EFA01652.1 NNAY01000079 OXU31180.1 ACPB03003277 KQ976681 KYM78186.1 GL888434 EGI60888.1 KZ288276 PBC29725.1 KK107151 EZA57376.1 GL445595 EFN89299.1 KQ435944 KOX68172.1 KQ979528 KYN21147.1 GBBI01002658 JAC16054.1 ADTU01004134 ADTU01004135 ADTU01004136 ADTU01004137 ADTU01004138 ADTU01004139 ADTU01004140 ADTU01004141 ADTU01004142 ADTU01004143 CH963857 EDW76568.1 CP012523 ALC39246.1 CH902620 EDV32148.1 KPU73816.1 KQ435037 KZC14143.1 GECU01036013 JAS71693.1 GEZM01057944 JAV72080.1 KQ982392 KYQ56881.1 CH933807 EDW11076.1 KRG02521.1 GEBQ01021963 JAT18014.1 KQ981276 KYN43739.1 QOIP01000003 RLU24413.1 CH916368 EDW03338.1 GDHF01011810 JAI40504.1 OUUW01000004 SPP79229.1 SPP79231.1 GEBQ01028211 JAT11766.1 CM000157 EDW87532.1 CH479180 EDW29073.1 CH954177 EDV57615.1 CH379060 EAL33829.2 KRT04224.1 CH480820 EDW54138.1 CM000361 CM002910 EDX03609.1 KMY87884.1 AE014134 BT010030 AY268064 KQ977696 KYN00571.1 CH940649 EDW63960.1 KQ414596 KOC70061.1 LBMM01004751 KMQ92109.1 KMY87886.1 GL437408 EFN70488.1 UFQT01000093 SSX19622.1 GBRD01007886 GBRD01001372 JAG57935.1 KK852777 KDR16775.1 GBYB01004676 JAG74443.1 KK854326 PTY14687.1 GBHO01019049 GDHC01016936 JAG24555.1 JAQ01693.1 GAMC01020672 JAB85883.1 GL768119 EFZ12159.1 GEDC01027945 GEDC01011111 JAS09353.1 JAS26187.1 GAKP01020467 JAC38485.1 KB632299 ERL91738.1 CCAG010020175 JXJN01020204 JRES01001205 KNC24622.1 GDHF01017963 JAI34351.1 GDHF01028060 JAI24254.1 GECU01017809 JAS89897.1 PYGN01000865 PSN39900.1 GDHF01003678 JAI48636.1 GDRN01058336 JAI65608.1 CH477726 EAT36839.1 DS235877 EEB19792.1 JXUM01107019 JXUM01107020 JXUM01107021 JXUM01107022 JXUM01107023 JXUM01107024 JXUM01107025 KQ565104 KXJ71333.1 DS232027 EDS32438.1 JTDY01003479 KOB69495.1 AAAB01008980 EAA14535.4 APCN01001629 AXCN02001979 GFDL01005446 JAV29599.1 AXCM01000301 GFDL01005438 JAV29607.1 NEDP02003995 OWF47223.1 ADMH02001289 ETN63135.1
Proteomes
UP000218220
UP000053268
UP000005204
UP000053240
UP000007266
UP000215335
+ More
UP000015103 UP000078540 UP000007755 UP000242457 UP000005203 UP000053097 UP000008237 UP000053105 UP000078492 UP000005205 UP000007798 UP000092553 UP000007801 UP000076502 UP000075809 UP000009192 UP000078541 UP000279307 UP000001070 UP000268350 UP000002282 UP000008744 UP000008711 UP000001819 UP000192221 UP000001292 UP000000304 UP000000803 UP000078542 UP000192223 UP000008792 UP000053825 UP000036403 UP000000311 UP000079169 UP000027135 UP000095301 UP000030742 UP000092444 UP000092445 UP000078200 UP000092443 UP000092460 UP000037069 UP000095300 UP000245037 UP000091820 UP000008820 UP000009046 UP000069940 UP000249989 UP000002320 UP000037510 UP000075903 UP000007062 UP000076407 UP000075885 UP000075840 UP000075901 UP000075902 UP000075886 UP000075881 UP000075883 UP000242188 UP000076408 UP000075884 UP000075900 UP000000673 UP000075920
UP000015103 UP000078540 UP000007755 UP000242457 UP000005203 UP000053097 UP000008237 UP000053105 UP000078492 UP000005205 UP000007798 UP000092553 UP000007801 UP000076502 UP000075809 UP000009192 UP000078541 UP000279307 UP000001070 UP000268350 UP000002282 UP000008744 UP000008711 UP000001819 UP000192221 UP000001292 UP000000304 UP000000803 UP000078542 UP000192223 UP000008792 UP000053825 UP000036403 UP000000311 UP000079169 UP000027135 UP000095301 UP000030742 UP000092444 UP000092445 UP000078200 UP000092443 UP000092460 UP000037069 UP000095300 UP000245037 UP000091820 UP000008820 UP000009046 UP000069940 UP000249989 UP000002320 UP000037510 UP000075903 UP000007062 UP000076407 UP000075885 UP000075840 UP000075901 UP000075902 UP000075886 UP000075881 UP000075883 UP000242188 UP000076408 UP000075884 UP000075900 UP000000673 UP000075920
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2A4JVU1
A0A2H1V7K7
A0A194PVS1
H9JYD6
A0A194REM4
I4DLJ5
+ More
D2A0Q4 A0A232FKY8 T1I445 A0A195B151 F4WY07 A0A2A3EDA0 A0A088ANE0 A0A026WNX2 E2B4S0 A0A0N0BC47 A0A151J8G2 A0A023F4X4 A0A158NZ99 B4MWS9 A0A0M4E5A5 B3MNW1 A0A154PQG7 A0A1B6HAJ5 A0A1Y1LIC7 A0A151X933 B4KG89 A0A1B6L3A7 A0A195FV45 A0A3L8DW00 B4JDG8 A0A0K8VNP3 A0A3B0JZV2 A0A3B0K2J7 A0A1B6KJZ6 B4NXW1 B4G9P3 B3N316 Q29M65 A0A1W4UTS1 B4I2Z3 B4Q9J8 Q6WV19 A0A195CIK7 A0A1W4X6X2 B4LTQ9 A0A0L7RH75 A0A0J7KPE7 A0A0J9QV41 E2A7V1 A0A1S4EP92 A0A336LNH0 A0A0K8SYF1 A0A067R1I4 A0A0C9RCP2 A0A2R7W4N3 A0A0A9Y4T6 W8AN19 E9J4V7 A0A1B6C795 A0A1I8NIG4 A0A034V9M4 U4UH93 A0A1B0G2M0 A0A1B0A0V0 A0A1A9VTU0 A0A1A9Y369 A0A1B0BTJ8 A0A0L0BX46 A0A0K8V659 A0A0K8UCA0 A0A1I8NZS1 A0A1B6ISM0 A0A2P8Y6L9 A0A0K8WBU9 A0A0N7ZCW8 A0A1A9W0R7 Q16R35 E0W2D6 A0A182H3A2 B0WPN5 A0A0L7L238 A0A182V2F4 Q7Q0X9 A0A182XH81 A0A182PTI8 A0A182HR03 A0A182SZN9 A0A182TV63 A0A182Q6H4 A0A182JVQ3 A0A1Q3FPR8 A0A182LSH7 A0A1Q3FPW6 A0A210QER2 A0A182YQN2 A0A182NNR3 A0A182R1P7 W5JJQ0 A0A182W8H1
D2A0Q4 A0A232FKY8 T1I445 A0A195B151 F4WY07 A0A2A3EDA0 A0A088ANE0 A0A026WNX2 E2B4S0 A0A0N0BC47 A0A151J8G2 A0A023F4X4 A0A158NZ99 B4MWS9 A0A0M4E5A5 B3MNW1 A0A154PQG7 A0A1B6HAJ5 A0A1Y1LIC7 A0A151X933 B4KG89 A0A1B6L3A7 A0A195FV45 A0A3L8DW00 B4JDG8 A0A0K8VNP3 A0A3B0JZV2 A0A3B0K2J7 A0A1B6KJZ6 B4NXW1 B4G9P3 B3N316 Q29M65 A0A1W4UTS1 B4I2Z3 B4Q9J8 Q6WV19 A0A195CIK7 A0A1W4X6X2 B4LTQ9 A0A0L7RH75 A0A0J7KPE7 A0A0J9QV41 E2A7V1 A0A1S4EP92 A0A336LNH0 A0A0K8SYF1 A0A067R1I4 A0A0C9RCP2 A0A2R7W4N3 A0A0A9Y4T6 W8AN19 E9J4V7 A0A1B6C795 A0A1I8NIG4 A0A034V9M4 U4UH93 A0A1B0G2M0 A0A1B0A0V0 A0A1A9VTU0 A0A1A9Y369 A0A1B0BTJ8 A0A0L0BX46 A0A0K8V659 A0A0K8UCA0 A0A1I8NZS1 A0A1B6ISM0 A0A2P8Y6L9 A0A0K8WBU9 A0A0N7ZCW8 A0A1A9W0R7 Q16R35 E0W2D6 A0A182H3A2 B0WPN5 A0A0L7L238 A0A182V2F4 Q7Q0X9 A0A182XH81 A0A182PTI8 A0A182HR03 A0A182SZN9 A0A182TV63 A0A182Q6H4 A0A182JVQ3 A0A1Q3FPR8 A0A182LSH7 A0A1Q3FPW6 A0A210QER2 A0A182YQN2 A0A182NNR3 A0A182R1P7 W5JJQ0 A0A182W8H1
PDB
2FFV
E-value=1.24926e-101,
Score=943
Ontologies
PATHWAY
GO
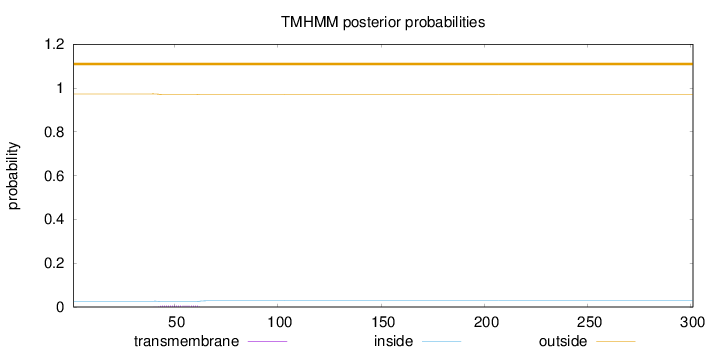
Topology
Subcellular location
Golgi apparatus membrane
Length:
301
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13556
Exp number, first 60 AAs:
0.12512
Total prob of N-in:
0.02676
outside
1 - 301
Population Genetic Test Statistics
Pi
22.088294
Theta
20.953933
Tajima's D
-1.069017
CLR
0.549251
CSRT
0.125643717814109
Interpretation
Uncertain
