Gene
KWMTBOMO07480
Pre Gene Modal
BGIBMGA005195
Annotation
PREDICTED:_serine/threonine-protein_phosphatase_6_regulatory_ankyrin_repeat_subunit_A_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 1.287
Sequence
CDS
ATGGTCCGAGAGCTGCTAAGCGCACAAACGGCTGAACAACTGAAGGCGACGACCCCTGCCGGAGACACCGCTCTGCATCTAGCTGCCAGAAGGAGGGACGTAGACATGGCCAGGATCTTAGTGGACTACGGAGCAGTAGTCGATGCTACGAATGGCGCTGGACAAACACCTTTACATATAGCCGCAGCTGAAGGCGATGAGCCCTTAGTAAAGTACTTCTACGGAGTCAGGGCTAACGCTGCGATTGCTGATAATGAGGATCGAACTCCAATGCATTTAGCAGCGGAGAATGGTCATGCTGCCATCATTGAACTACTGGCGGACAAGTTTAAGGCTTCGATCTTCGAGAGGACCAAGGACGGCAGCACTTTGATGCACATTGCGTCACTCAATGGCCACGCGGATTGCGCCATGATGCTTTTCAAGAAAGGTGTCTACCTACACATGCCCAACAAGGATGGAGCGCGGAGTATCCACACAGCGGCTCGTTATGGCCATGTTGGGATCATCAATACGTTGTTGCAGAAAGGGGAAAGTGTTGATGTCACGACCAATGATAACTACACAGCTCTCCATATTGCAGTTGAGTCGTGTAAACCAGCAGTGGTAGAAACTCTTTTAGGGTATGGCGCAGACGTTCATATTCGAGGTGGAAAACAAAGAGAAACTCCTCTCCACATAGCTGCTAGGATACCTGATGGAGACAAATGCGCGCTGATGTTGCTAAAGTCCGGGGCTGGCCCAAACAAGGCGACAGAAGATGGCATGACGCCTGTACATGTCGCCGCCAAATTTGGAAATCTGGCAACGCTGGTCTTACTACTGGAAGACGGTGGAGACCCTTTGCGGAAAACTAAAACAGGAGAAACGCCTTTACAAATGGCCTGTCGAAGCTGCATGCCAGATATTGTGCGACACTTAATAGAATTCGTAAAAGACCACAAAGGAGAAAGCGTGTCTACGGCATTTATAGACGCAGTAGACGAGGATGGAGCCAGCGCATTGCATTACGCTTGTAAAGTTACCAAAGAAGAAGTTAAAATACCTACAGCAGATAGACAGGTTGTCAAATGCTTAATAGAAAACGGCGCAGATGTGTCTCTGCAAACAAGGCATAACCACGAAACGGCTTTTCATTATTGCGCTATTGCTGGTAATAATGACGTCATGACTGAAATGATCGCGCATATCTCAACTGCGGACGTCAGCAGAGCGCTCAATAGACAAAATTCAATAGGGTGGACGCCTCTTCTAATAGCCTGTCACAGAGGACATATGCAACTCGTAAATACATTATTAACAAACCACGCCCGAGTTGACGTATTCGACGTCGAAGGCAGATCCGCATTGCATTTGGCAGCTGAGCACGGATACCTACAAGTATGTGACGCACTACTAACAAATAAAGCTTTCATAAATTCAAAAGCACGAAACGGCAGAACGGCTCTTCATTTGGCAGCAATGAACGGATACGCACATCTAGTCAAGTTTCTTATTAGAGACCACAACGCTATGATAGACGTTCTAACCTTGAAAAAGCAAACTCCACTACATTTGGCAGCTGCTAGTGGTCAGATCGAAGTATGTAAGCTTCTATTAGAACTTGGTGCGAATATTGATGCGACCGATGAGCTCGGACAAAAACCGATACACGCAGCTGCACAAAATAACTATTCAGAAGTCGTTCAATTGTTTTTACAACAGCACCCCAACTTGGTAATGGCAACAACTAAAGATGGAAACACGTGCGCTCATATTGCAGCCATACAAGGCTCTGTTAAAGTGATCGAGGAATTAATGAAGTTCGATAGGACTGGAGTTATTTCAGCTCGAAATAAACTGAATGATTCAACGCCTTTGCAACTAGCAGCGGAAGGTGGTCATGCAGATGTTGTAAGGGTGCTTGTGAGAGCCGGAGCATCGTGTACTGATGAAAACAGAGCCGGCCTCACTGCTGTACACTTGGCAGCAGAGCATGGACACACTAATGTATTGGATGTTATGAGATCAACGAATACTTTAAGGATATCTAGCAAAAAGTTGGGATTGACTCCATTACATATTGCAGCGTATTATGGACAAGCTGAAACTGTAAGAGAACTTCTATCTCACGTTCCCGGTACTGTGAAATCTGAGGCTCCTACGGGAGTCTCATTGGTCCCAGTATTAGGAGCTGAATCTGGTTTGACACCTCTCCATTTGGCTGCATATAACGGTAACGAGAACGTGGTCCGATTACTCCTCAACTCTGCCGGTGTCCAAGCGGATGCAGCTAGTAACGAAAATGGCTATAATCCATTACATCTAGCATGTTTCGGAGGCCATATGTCCATAGTAGGACTTCTATTAAGTAGATCAGCAGAACTACTACACAGTACTGACAGGCACGGAAAAACTGGTTTACACATTGCATCAACACACGGACATTATCAAATGGTTGAGGTCTTGTTGGGTCAAGGAGCAGAAATAAACGCTACCGATAAAAACGGATGGACACCATTGCACTGTGCAGCAAAAGCTGGTCACTTGAACGTCGTGAAACTACTTTGTGAATCCGGAGCATCTCCAAAAAGTGAGACCAATCTAAATTGCGCCCCTATATGGTTCGCCGCTTCGGAAAACCACAACGACGTTCTTGAATATTTACTCCACAAAGAACACGATACTCAGTCACTGATGGACGACAAGAGGTTTGTCTACAACCTCATGGTTTGTTCTAAGAACCACAATAACATTCCTATAGAGGAATTCGTTTTGGTATCACCGGCGCCGGTCGACACGGCTGCCAAGTTATCGAATATTTACATCAATTTATCGACTAAGGAAAAGGAGCGTGCCAAAGACCTTATAGCGGCAGGGAAACAATGTGAGGCGATGGCGACCGAACTGCTGGCTCTAGCGGCGGGGGCCGACTCTGCCGGACACATACTCACTGCCACGGATAACCGAAACATTGAATTCTTGGATGTTCTAATAGAAAACGAACAAAAAGAAGTAATCGCCCACACAGTTGTCCAGAGATATCTTCAGGAACTCTGGAGAGGGAGTCTCAAATGGACCGGCATCAAAATTATGTTCCTTTTTTTCGCATTTATAGTCTGTCCACCAGTTTGGATGGTGTTCTCTCTTCCCTTAGCTTGTGTCCAAATTTTGGACTTCTTATCCTTCCATCATCTGTTCGGTCCATGGGCGATAATTATAGGAGATCTCATGAAAGATTTGGGCAGATTTTTAGCAGTTTTAGCGATTTTTGTATTTGGATTCTCGATGCATATAGTAGCATTGAATCAGCCATTCAGAAATATTAATAAAAGTGAAGACAATAAGTATGCAAGCCAAGCCAGAAGAAAACTTTTCTCAGACGTCACAATGAATCCGCTGTTTTCGTTCGAGCTACTCTTCTTTGCGGTTTTCGGGCAGACAACTACAGAACAAACAAAGGTTCATAGTAAGGATAGTAACATCCAGCCCGCTTGGACGAATTACTTATTCAAGATAGTCTTTGGGATCTATATGCTGGTGTCAGTAGTGGTTCTCATCAACCTACTGATCGCTATGATGTCAGACACCTACCAGCGGATACAGGCGCAATCCGACATCGAATGGAAGTATGGCTTGTCAAAGTTGATAAGAAATATGCACAGAACGAATACGGCTCCGTCGCCCCTCAACTTGGTCACGACGTGGCTTATGTGGTTGATTGCCAGATGTCGAGAACGGCTTACCAAAAAGAAGAGGCCAAGCTTAGTACACATGATTGGCTTGCAGAGGCAAGATCAGTTGAGTGCGAGATCGAAGGCCGGTGCTAAGTGGCTATCCAAAGTCAAGAGAGGACAAGTTGTACCTAAAGACTCGACGAGACTATCAGTAGTCCACTTAAGTCCTCTAGGTTCGCAGCTCAGTTTCAACAACGCAACGAGGATAGAAAACGTAGTAGACTGGGAGATTATAGCGAAGAAGTATAGAGCTCTGATGAGGGATGAGCCTGAGGAGTCCACAGCAAAGGAAAGCGAAACGGACTCATCTGATGAAGTATCAGAAATTATCCCAAATAACATAGCCGCAGCTCCGCCTTGA
Protein
MVRELLSAQTAEQLKATTPAGDTALHLAARRRDVDMARILVDYGAVVDATNGAGQTPLHIAAAEGDEPLVKYFYGVRANAAIADNEDRTPMHLAAENGHAAIIELLADKFKASIFERTKDGSTLMHIASLNGHADCAMMLFKKGVYLHMPNKDGARSIHTAARYGHVGIINTLLQKGESVDVTTNDNYTALHIAVESCKPAVVETLLGYGADVHIRGGKQRETPLHIAARIPDGDKCALMLLKSGAGPNKATEDGMTPVHVAAKFGNLATLVLLLEDGGDPLRKTKTGETPLQMACRSCMPDIVRHLIEFVKDHKGESVSTAFIDAVDEDGASALHYACKVTKEEVKIPTADRQVVKCLIENGADVSLQTRHNHETAFHYCAIAGNNDVMTEMIAHISTADVSRALNRQNSIGWTPLLIACHRGHMQLVNTLLTNHARVDVFDVEGRSALHLAAEHGYLQVCDALLTNKAFINSKARNGRTALHLAAMNGYAHLVKFLIRDHNAMIDVLTLKKQTPLHLAAASGQIEVCKLLLELGANIDATDELGQKPIHAAAQNNYSEVVQLFLQQHPNLVMATTKDGNTCAHIAAIQGSVKVIEELMKFDRTGVISARNKLNDSTPLQLAAEGGHADVVRVLVRAGASCTDENRAGLTAVHLAAEHGHTNVLDVMRSTNTLRISSKKLGLTPLHIAAYYGQAETVRELLSHVPGTVKSEAPTGVSLVPVLGAESGLTPLHLAAYNGNENVVRLLLNSAGVQADAASNENGYNPLHLACFGGHMSIVGLLLSRSAELLHSTDRHGKTGLHIASTHGHYQMVEVLLGQGAEINATDKNGWTPLHCAAKAGHLNVVKLLCESGASPKSETNLNCAPIWFAASENHNDVLEYLLHKEHDTQSLMDDKRFVYNLMVCSKNHNNIPIEEFVLVSPAPVDTAAKLSNIYINLSTKEKERAKDLIAAGKQCEAMATELLALAAGADSAGHILTATDNRNIEFLDVLIENEQKEVIAHTVVQRYLQELWRGSLKWTGIKIMFLFFAFIVCPPVWMVFSLPLACVQILDFLSFHHLFGPWAIIIGDLMKDLGRFLAVLAIFVFGFSMHIVALNQPFRNINKSEDNKYASQARRKLFSDVTMNPLFSFELLFFAVFGQTTTEQTKVHSKDSNIQPAWTNYLFKIVFGIYMLVSVVVLINLLIAMMSDTYQRIQAQSDIEWKYGLSKLIRNMHRTNTAPSPLNLVTTWLMWLIARCRERLTKKKRPSLVHMIGLQRQDQLSARSKAGAKWLSKVKRGQVVPKDSTRLSVVHLSPLGSQLSFNNATRIENVVDWEIIAKKYRALMRDEPEESTAKESETDSSDEVSEIIPNNIAAAPP
Summary
Similarity
Belongs to the transient receptor (TC 1.A.4) family.
Uniprot
A0A2A4JNI4
A0A2H1VHA0
A0A3S2NYX8
A0A194PTV6
A0A212F919
A0A194REK9
+ More
A0A0L0CG13 A0A1I8MTT7 A0A1I8MTR8 A0A1I8Q5K0 A0A1I8Q5F7 A0A0Q9XCZ4 A0A0Q9X899 B4KF23 B4LU24 B4MVL1 B4JAG5 B5DHR9 A0A0R3NXU0 A0A3B0KF06 A0A0R3NX30 B4GJM6 Q7KTN8 E0A9E1 A0A0J9TG65 A0A0R3NRT7 A0A0R3NRK2 B4P068 A0A0R1DL79 A0A1W4UVZ3 A0A0J9QW54 A0A0R3NWQ2 A0A0R1DSQ1 A0A1W4UVZ8 A0A1W4UIZ2 A0A1W4UW89 A0A0R3NRG1 A8DYV6 A0A0R3NRC1 A0A0Q5W9L1 A0A1B0A4D2 A0A0J9QW80 Q9VMR4 A0A0S0WN65 Q7KIQ2 A0A1W4UXN3 A0A1W4UIF7 A0A1B0FIS4 A0A1A9V813 A0A0Q5W9Z0 A0A0R3NYM8 A0A0R1DLI7 B3N4V4 B3MLL2 A0A0J9QX51 A0A0P8XFR4 A0A0J9TG59 A0A0R3NRB8 A0A0M3QTB0 A0A0R3NRL2 A0A0J9QW73 A0A0P9A519 Q16PD7 A0A1B0CH34 A0A1B0AYW3 A0A182QSQ1 A0A182Y9Q2 A0A1A9WXR4 A0A1S4GZD0 A0A1S4GZB8 A0A154PDA1 Q7Q8L0 A0A182NT94 A0A182KMJ6 A0A182VHG8 A0A182JI68 A0A182LT19 A0A182F1T1 A0A182P4L5 A0A084VUX0 A0A182VSF7 E2A4Y4 B0W065 A0A310SMG0 A0A195DFF2 A0A195FU27 A0A087ZYV2 A0A0T6AZN1 A0A158NYK8 A0A182WRU6 A0A1J1IM79 A0A182IBQ8 A0A139WC49 A0A139WBZ3 F4WY25 A0A182RWB4 A0A182TNL7 A0A0Q5WL74 A0A151IKH1
A0A0L0CG13 A0A1I8MTT7 A0A1I8MTR8 A0A1I8Q5K0 A0A1I8Q5F7 A0A0Q9XCZ4 A0A0Q9X899 B4KF23 B4LU24 B4MVL1 B4JAG5 B5DHR9 A0A0R3NXU0 A0A3B0KF06 A0A0R3NX30 B4GJM6 Q7KTN8 E0A9E1 A0A0J9TG65 A0A0R3NRT7 A0A0R3NRK2 B4P068 A0A0R1DL79 A0A1W4UVZ3 A0A0J9QW54 A0A0R3NWQ2 A0A0R1DSQ1 A0A1W4UVZ8 A0A1W4UIZ2 A0A1W4UW89 A0A0R3NRG1 A8DYV6 A0A0R3NRC1 A0A0Q5W9L1 A0A1B0A4D2 A0A0J9QW80 Q9VMR4 A0A0S0WN65 Q7KIQ2 A0A1W4UXN3 A0A1W4UIF7 A0A1B0FIS4 A0A1A9V813 A0A0Q5W9Z0 A0A0R3NYM8 A0A0R1DLI7 B3N4V4 B3MLL2 A0A0J9QX51 A0A0P8XFR4 A0A0J9TG59 A0A0R3NRB8 A0A0M3QTB0 A0A0R3NRL2 A0A0J9QW73 A0A0P9A519 Q16PD7 A0A1B0CH34 A0A1B0AYW3 A0A182QSQ1 A0A182Y9Q2 A0A1A9WXR4 A0A1S4GZD0 A0A1S4GZB8 A0A154PDA1 Q7Q8L0 A0A182NT94 A0A182KMJ6 A0A182VHG8 A0A182JI68 A0A182LT19 A0A182F1T1 A0A182P4L5 A0A084VUX0 A0A182VSF7 E2A4Y4 B0W065 A0A310SMG0 A0A195DFF2 A0A195FU27 A0A087ZYV2 A0A0T6AZN1 A0A158NYK8 A0A182WRU6 A0A1J1IM79 A0A182IBQ8 A0A139WC49 A0A139WBZ3 F4WY25 A0A182RWB4 A0A182TNL7 A0A0Q5WL74 A0A151IKH1
Pubmed
EMBL
NWSH01000898
PCG73635.1
ODYU01002554
SOQ40210.1
RSAL01000020
RVE52613.1
+ More
KQ459592 KPI96846.1 AGBW02009654 OWR50221.1 KQ460325 KPJ15725.1 JRES01000438 KNC31186.1 CH933807 KRG03546.1 KRG03545.1 EDW13006.1 CH940649 EDW65077.1 CH963857 EDW75731.2 CH916368 EDW03836.1 CH379058 EDY69846.2 KRT03635.1 OUUW01000006 SPP82188.1 KRT03637.1 CH479184 EDW36842.1 AE014134 AAS64642.2 HM582118 ADK73985.1 AFH03565.1 CM002910 KMY88295.1 KRT03644.1 KRT03641.1 CM000157 EDW88933.2 KRJ98107.1 KMY88292.1 KRT03645.1 KRJ98105.1 KRT03643.1 ABV53627.2 KRT03634.1 KRT03636.1 KRT03640.1 CH954177 KQS70179.1 KMY88293.1 AAF52248.4 ALI30195.1 AF242296 AAF59842.1 CCAG010004544 KQS70180.1 KRT03638.1 KRJ98106.1 EDV57856.2 CH902620 EDV31761.2 KMY88294.1 KPU73610.1 KMY88290.1 KRT03639.1 CP012523 ALC38569.1 KRT03642.1 KMY88291.1 KPU73611.1 CH477786 EAT36228.1 AJWK01011958 AJWK01011959 AJWK01011960 AJWK01011961 AJWK01011962 AJWK01011963 AJWK01011964 JXJN01006000 JXJN01006001 JXJN01006002 JXJN01006003 JXJN01006004 JXJN01006005 AXCN02000723 AAAB01008944 KQ434875 KZC09787.1 EAA10081.4 AXCM01004451 ATLV01017052 ATLV01017053 KE525143 KFB41764.1 GL436770 EFN71501.1 DS231816 EDS37637.1 KQ759870 OAD62435.1 KQ980953 KYN11179.1 KQ981276 KYN43797.1 LJIG01022446 KRT80566.1 ADTU01004019 ADTU01004020 CVRI01000054 CRL00842.1 APCN01000724 KQ971371 KYB25487.1 KYB25488.1 GL888434 EGI60950.1 KQS70178.1 KQ977202 KYN04979.1
KQ459592 KPI96846.1 AGBW02009654 OWR50221.1 KQ460325 KPJ15725.1 JRES01000438 KNC31186.1 CH933807 KRG03546.1 KRG03545.1 EDW13006.1 CH940649 EDW65077.1 CH963857 EDW75731.2 CH916368 EDW03836.1 CH379058 EDY69846.2 KRT03635.1 OUUW01000006 SPP82188.1 KRT03637.1 CH479184 EDW36842.1 AE014134 AAS64642.2 HM582118 ADK73985.1 AFH03565.1 CM002910 KMY88295.1 KRT03644.1 KRT03641.1 CM000157 EDW88933.2 KRJ98107.1 KMY88292.1 KRT03645.1 KRJ98105.1 KRT03643.1 ABV53627.2 KRT03634.1 KRT03636.1 KRT03640.1 CH954177 KQS70179.1 KMY88293.1 AAF52248.4 ALI30195.1 AF242296 AAF59842.1 CCAG010004544 KQS70180.1 KRT03638.1 KRJ98106.1 EDV57856.2 CH902620 EDV31761.2 KMY88294.1 KPU73610.1 KMY88290.1 KRT03639.1 CP012523 ALC38569.1 KRT03642.1 KMY88291.1 KPU73611.1 CH477786 EAT36228.1 AJWK01011958 AJWK01011959 AJWK01011960 AJWK01011961 AJWK01011962 AJWK01011963 AJWK01011964 JXJN01006000 JXJN01006001 JXJN01006002 JXJN01006003 JXJN01006004 JXJN01006005 AXCN02000723 AAAB01008944 KQ434875 KZC09787.1 EAA10081.4 AXCM01004451 ATLV01017052 ATLV01017053 KE525143 KFB41764.1 GL436770 EFN71501.1 DS231816 EDS37637.1 KQ759870 OAD62435.1 KQ980953 KYN11179.1 KQ981276 KYN43797.1 LJIG01022446 KRT80566.1 ADTU01004019 ADTU01004020 CVRI01000054 CRL00842.1 APCN01000724 KQ971371 KYB25487.1 KYB25488.1 GL888434 EGI60950.1 KQS70178.1 KQ977202 KYN04979.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000007151
UP000053240
UP000037069
+ More
UP000095301 UP000095300 UP000009192 UP000008792 UP000007798 UP000001070 UP000001819 UP000268350 UP000008744 UP000000803 UP000002282 UP000192221 UP000008711 UP000092445 UP000092444 UP000078200 UP000007801 UP000092553 UP000008820 UP000092461 UP000092460 UP000075886 UP000076408 UP000091820 UP000076502 UP000007062 UP000075884 UP000075882 UP000075903 UP000075880 UP000075883 UP000069272 UP000075885 UP000030765 UP000075920 UP000000311 UP000002320 UP000078492 UP000078541 UP000005203 UP000005205 UP000076407 UP000183832 UP000075840 UP000007266 UP000007755 UP000075900 UP000075902 UP000078542
UP000095301 UP000095300 UP000009192 UP000008792 UP000007798 UP000001070 UP000001819 UP000268350 UP000008744 UP000000803 UP000002282 UP000192221 UP000008711 UP000092445 UP000092444 UP000078200 UP000007801 UP000092553 UP000008820 UP000092461 UP000092460 UP000075886 UP000076408 UP000091820 UP000076502 UP000007062 UP000075884 UP000075882 UP000075903 UP000075880 UP000075883 UP000069272 UP000075885 UP000030765 UP000075920 UP000000311 UP000002320 UP000078492 UP000078541 UP000005203 UP000005205 UP000076407 UP000183832 UP000075840 UP000007266 UP000007755 UP000075900 UP000075902 UP000078542
Interpro
SUPFAM
SSF48403
SSF48403
Gene 3D
CDD
ProteinModelPortal
A0A2A4JNI4
A0A2H1VHA0
A0A3S2NYX8
A0A194PTV6
A0A212F919
A0A194REK9
+ More
A0A0L0CG13 A0A1I8MTT7 A0A1I8MTR8 A0A1I8Q5K0 A0A1I8Q5F7 A0A0Q9XCZ4 A0A0Q9X899 B4KF23 B4LU24 B4MVL1 B4JAG5 B5DHR9 A0A0R3NXU0 A0A3B0KF06 A0A0R3NX30 B4GJM6 Q7KTN8 E0A9E1 A0A0J9TG65 A0A0R3NRT7 A0A0R3NRK2 B4P068 A0A0R1DL79 A0A1W4UVZ3 A0A0J9QW54 A0A0R3NWQ2 A0A0R1DSQ1 A0A1W4UVZ8 A0A1W4UIZ2 A0A1W4UW89 A0A0R3NRG1 A8DYV6 A0A0R3NRC1 A0A0Q5W9L1 A0A1B0A4D2 A0A0J9QW80 Q9VMR4 A0A0S0WN65 Q7KIQ2 A0A1W4UXN3 A0A1W4UIF7 A0A1B0FIS4 A0A1A9V813 A0A0Q5W9Z0 A0A0R3NYM8 A0A0R1DLI7 B3N4V4 B3MLL2 A0A0J9QX51 A0A0P8XFR4 A0A0J9TG59 A0A0R3NRB8 A0A0M3QTB0 A0A0R3NRL2 A0A0J9QW73 A0A0P9A519 Q16PD7 A0A1B0CH34 A0A1B0AYW3 A0A182QSQ1 A0A182Y9Q2 A0A1A9WXR4 A0A1S4GZD0 A0A1S4GZB8 A0A154PDA1 Q7Q8L0 A0A182NT94 A0A182KMJ6 A0A182VHG8 A0A182JI68 A0A182LT19 A0A182F1T1 A0A182P4L5 A0A084VUX0 A0A182VSF7 E2A4Y4 B0W065 A0A310SMG0 A0A195DFF2 A0A195FU27 A0A087ZYV2 A0A0T6AZN1 A0A158NYK8 A0A182WRU6 A0A1J1IM79 A0A182IBQ8 A0A139WC49 A0A139WBZ3 F4WY25 A0A182RWB4 A0A182TNL7 A0A0Q5WL74 A0A151IKH1
A0A0L0CG13 A0A1I8MTT7 A0A1I8MTR8 A0A1I8Q5K0 A0A1I8Q5F7 A0A0Q9XCZ4 A0A0Q9X899 B4KF23 B4LU24 B4MVL1 B4JAG5 B5DHR9 A0A0R3NXU0 A0A3B0KF06 A0A0R3NX30 B4GJM6 Q7KTN8 E0A9E1 A0A0J9TG65 A0A0R3NRT7 A0A0R3NRK2 B4P068 A0A0R1DL79 A0A1W4UVZ3 A0A0J9QW54 A0A0R3NWQ2 A0A0R1DSQ1 A0A1W4UVZ8 A0A1W4UIZ2 A0A1W4UW89 A0A0R3NRG1 A8DYV6 A0A0R3NRC1 A0A0Q5W9L1 A0A1B0A4D2 A0A0J9QW80 Q9VMR4 A0A0S0WN65 Q7KIQ2 A0A1W4UXN3 A0A1W4UIF7 A0A1B0FIS4 A0A1A9V813 A0A0Q5W9Z0 A0A0R3NYM8 A0A0R1DLI7 B3N4V4 B3MLL2 A0A0J9QX51 A0A0P8XFR4 A0A0J9TG59 A0A0R3NRB8 A0A0M3QTB0 A0A0R3NRL2 A0A0J9QW73 A0A0P9A519 Q16PD7 A0A1B0CH34 A0A1B0AYW3 A0A182QSQ1 A0A182Y9Q2 A0A1A9WXR4 A0A1S4GZD0 A0A1S4GZB8 A0A154PDA1 Q7Q8L0 A0A182NT94 A0A182KMJ6 A0A182VHG8 A0A182JI68 A0A182LT19 A0A182F1T1 A0A182P4L5 A0A084VUX0 A0A182VSF7 E2A4Y4 B0W065 A0A310SMG0 A0A195DFF2 A0A195FU27 A0A087ZYV2 A0A0T6AZN1 A0A158NYK8 A0A182WRU6 A0A1J1IM79 A0A182IBQ8 A0A139WC49 A0A139WBZ3 F4WY25 A0A182RWB4 A0A182TNL7 A0A0Q5WL74 A0A151IKH1
Ontologies
GO
GO:0005262
GO:0016021
GO:0006811
GO:0005216
GO:0010996
GO:0030425
GO:0040011
GO:0071260
GO:0007605
GO:0001964
GO:0050976
GO:0007638
GO:0043025
GO:0005929
GO:0044214
GO:0030506
GO:0050954
GO:0006816
GO:0006812
GO:0034703
GO:0005261
GO:0050975
GO:0050974
GO:0008092
GO:0008381
GO:0005515
GO:0016020
GO:0005509
GO:0016042
GO:0003707
GO:0051537
GO:0055085
GO:0070588
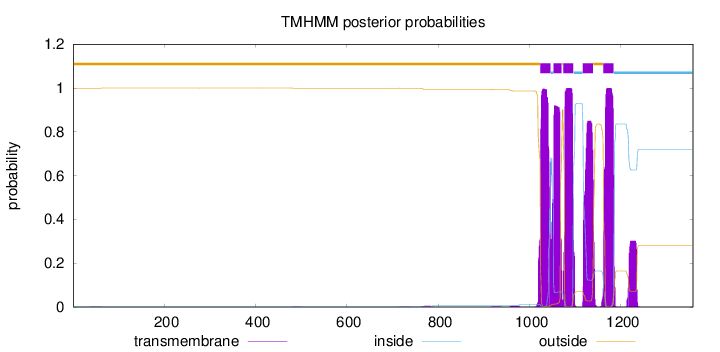
Topology
Length:
1358
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
106.48217
Exp number, first 60 AAs:
0.0059
Total prob of N-in:
0.00057
outside
1 - 1023
TMhelix
1024 - 1046
inside
1047 - 1052
TMhelix
1053 - 1070
outside
1071 - 1073
TMhelix
1074 - 1096
inside
1097 - 1116
TMhelix
1117 - 1139
outside
1140 - 1161
TMhelix
1162 - 1184
inside
1185 - 1358
Population Genetic Test Statistics
Pi
20.265302
Theta
19.138046
Tajima's D
0.407669
CLR
0.976727
CSRT
0.496625168741563
Interpretation
Uncertain
