Gene
KWMTBOMO07459
Pre Gene Modal
BGIBMGA005187
Annotation
PREDICTED:_pancreatic_triacylglycerol_lipase-like_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 1.817
Sequence
CDS
ATGTTAGACTGGGGCTCGCTTTGTCAGCCTCCGTGCTACGTCGCAGCGGTCCACAATCTGAGACCTGTTGCTCGATGCGCGGCTGAGGCACTGGGCACTCTGAGACGCGCCGGCTTGAGGCCAGACCGATTAACATGCGTGGGACACTCGTTGGGTGCACATATGTGTGGCATCATTGCTAACTTCCTTACGTTCCGGCTAAACCGAATTATAGGGTTGGATCCAGCCCGACCTTTAATTCGGTCCGCCCCTGCCCTACGATTGGACCCAGGGGATGCGCGCGCCGTCCACGTACTCCACACCAACGCAGGTCGCTATGGTGAAGGTGCCAGGCTTGGACACGCTGACTTCTGTCTTAACGGGGGACGCAGTCAACCATACTGCGAGGATACGCCAAACGAAGCGTTATGCAGCCATATCTGGTCAGTCTGCTATCAGGCGGAAAGTCTGTTCCGTTCTCGTAGCGCAGTTCCATGCGGCCGGCGTTGTTCAGTCCGCGTCCCGCCAGGACGTGCGCAAGCTCTGCCCGTGCCGGTCGGGCAGCCTGCGCCCATGACTGCATCAGGCGCATACTGTTTAGAAGACGACACTATACCGTTCTGCCCTCACACGTCGGGACTGTTGGAAGGTGACACACGCTGTTGCCTAGACGAGCAGTACGCAGCCGCGGCCACTAGCCCGCCGGCGCCGGTACGGAAGCGACCACGCCCCATCGTCAGGCTTAGAGCGAGCAGAGTCAAACTAGACGTCCACGATACTTTAGAATGA
Protein
MLDWGSLCQPPCYVAAVHNLRPVARCAAEALGTLRRAGLRPDRLTCVGHSLGAHMCGIIANFLTFRLNRIIGLDPARPLIRSAPALRLDPGDARAVHVLHTNAGRYGEGARLGHADFCLNGGRSQPYCEDTPNEALCSHIWSVCYQAESLFRSRSAVPCGRRCSVRVPPGRAQALPVPVGQPAPMTASGAYCLEDDTIPFCPHTSGLLEGDTRCCLDEQYAAAATSPPAPVRKRPRPIVRLRASRVKLDVHDTLE
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
H9J6P5
A0A2H1VRN5
A0A0N1PHU7
A0A194Q079
A0A212FF39
A0A3S2M6M2
+ More
A0A0K8TL87 A0A336K9X1 A0A182JD12 A0A182QP58 A0A2C9H8Q9 A0A1S4H056 A0A182V5D4 A0A182KUQ3 A0A1J1IWQ7 Q8ITU8 A0A182MBT5 A0A1S4FFD6 Q173Z4 A0A182GWJ6 A0A182U9H5 A0A2C9GRV7 A0A182F5H1 D6X109 A0A0M9A1C7 A0A084VCS5 A0A182K8P8 A0A2A3EU98 W5J9S5 A0A182R9Q1 A0A154P6W0 A0A088AGW0 A0A0L7QWJ1 A0A0C9PQR6 A0A0L7LNJ9 A0A0C9QJ60 T1GFZ3 A0A182YFE9 A0A232F8T8 N6SZV3 U4TTN9 A0A182PDD5 K7IRS5 U4UK56 A0A1B6KK83 T1H875 E9IG57 A0A151WNE0 A0A158NQR1 A0A195BEA4 F4WBU9 A0A195DI41 A0A195FSN8 J9K567 A0A2S2NWL8 A0A195D3J4 A0A067RF91 A0A2S2QFF1 E1ZX99 A0A2H8TH74 A0A3L8E398 A0A026WLQ1 A0A182WGZ1 A0A0A9WQ61 A0A0K8TB47 A0A1B6EGK5 A0A2J7R8E4 A0A084VCS6 A0A182PDD4 A0A182TB02 A0A182R9Q0 A0A182YFE8 A0A182MMH0 A0A182QMI5 A0A2C9H8S2 A0A2C9GS68 T1GFZ4 A0A1A9UMK0 A0A0T6BFB5 A0A0L0BZD9 E0VRR9 A0A1A9XZJ2 T1H8M0 A0A1W4WZZ2 Q173Z3 A0A1A9WZW6 A0A2S2Q5X0 A0A2R7WM19 A0A1S4FFC6 A0A2S2PDY1
A0A0K8TL87 A0A336K9X1 A0A182JD12 A0A182QP58 A0A2C9H8Q9 A0A1S4H056 A0A182V5D4 A0A182KUQ3 A0A1J1IWQ7 Q8ITU8 A0A182MBT5 A0A1S4FFD6 Q173Z4 A0A182GWJ6 A0A182U9H5 A0A2C9GRV7 A0A182F5H1 D6X109 A0A0M9A1C7 A0A084VCS5 A0A182K8P8 A0A2A3EU98 W5J9S5 A0A182R9Q1 A0A154P6W0 A0A088AGW0 A0A0L7QWJ1 A0A0C9PQR6 A0A0L7LNJ9 A0A0C9QJ60 T1GFZ3 A0A182YFE9 A0A232F8T8 N6SZV3 U4TTN9 A0A182PDD5 K7IRS5 U4UK56 A0A1B6KK83 T1H875 E9IG57 A0A151WNE0 A0A158NQR1 A0A195BEA4 F4WBU9 A0A195DI41 A0A195FSN8 J9K567 A0A2S2NWL8 A0A195D3J4 A0A067RF91 A0A2S2QFF1 E1ZX99 A0A2H8TH74 A0A3L8E398 A0A026WLQ1 A0A182WGZ1 A0A0A9WQ61 A0A0K8TB47 A0A1B6EGK5 A0A2J7R8E4 A0A084VCS6 A0A182PDD4 A0A182TB02 A0A182R9Q0 A0A182YFE8 A0A182MMH0 A0A182QMI5 A0A2C9H8S2 A0A2C9GS68 T1GFZ4 A0A1A9UMK0 A0A0T6BFB5 A0A0L0BZD9 E0VRR9 A0A1A9XZJ2 T1H8M0 A0A1W4WZZ2 Q173Z3 A0A1A9WZW6 A0A2S2Q5X0 A0A2R7WM19 A0A1S4FFC6 A0A2S2PDY1
Pubmed
EMBL
BABH01038149
BABH01038150
ODYU01004038
SOQ43511.1
KQ460636
KPJ13388.1
+ More
KQ459592 KPI96820.1 AGBW02008864 OWR52338.1 RSAL01000020 RVE52624.1 GDAI01002491 JAI15112.1 UFQS01000230 UFQT01000230 SSX01714.1 SSX22094.1 AXCN02000839 AAAB01008984 CVRI01000061 CRL03996.1 AF303984 AAN31169.1 AXCM01005154 CH477415 EAT41376.1 JXUM01093479 JXUM01093480 JXUM01093481 JXUM01093482 KQ564059 KXJ72856.1 APCN01003737 KQ971371 EFA09985.1 KQ435794 KOX73893.1 ATLV01010846 ATLV01010847 KE524620 KFB35769.1 KZ288189 PBC34631.1 ADMH02001866 ETN60751.1 KQ434827 KZC07602.1 KQ414713 KOC62970.1 GBYB01003583 JAG73350.1 JTDY01000493 KOB76934.1 GBYB01003584 JAG73351.1 CAQQ02088132 NNAY01000701 OXU26910.1 APGK01050307 KB741169 ENN73324.1 KB629940 ERL83298.1 KB632336 ERL92843.1 GEBQ01028109 JAT11868.1 ACPB03014203 ACPB03014204 ACPB03014205 GL762910 EFZ20475.1 KQ982911 KYQ49328.1 ADTU01023471 KQ976511 KYM82527.1 GL888066 EGI68377.1 KQ980824 KYN12563.1 KQ981280 KYN43461.1 ABLF02039488 GGMR01009004 MBY21623.1 KQ976885 KYN07473.1 KK852543 KDR21673.1 GGMS01007253 MBY76456.1 GL435030 EFN74189.1 GFXV01001671 MBW13476.1 QOIP01000001 RLU27012.1 KK107154 EZA56883.1 GBHO01033665 GBRD01003088 JAG09939.1 JAG62733.1 GBRD01003087 JAG62734.1 GEDC01000231 JAS37067.1 NEVH01006721 PNF37113.1 KFB35770.1 CAQQ02088126 CAQQ02088127 CAQQ02088128 CAQQ02088129 LJIG01000941 KRT85999.1 JRES01001119 KNC25385.1 DS235494 EEB16075.1 ACPB03007015 EAT41377.1 GGMS01003952 MBY73155.1 KK855069 PTY20664.1 GGMR01014986 MBY27605.1
KQ459592 KPI96820.1 AGBW02008864 OWR52338.1 RSAL01000020 RVE52624.1 GDAI01002491 JAI15112.1 UFQS01000230 UFQT01000230 SSX01714.1 SSX22094.1 AXCN02000839 AAAB01008984 CVRI01000061 CRL03996.1 AF303984 AAN31169.1 AXCM01005154 CH477415 EAT41376.1 JXUM01093479 JXUM01093480 JXUM01093481 JXUM01093482 KQ564059 KXJ72856.1 APCN01003737 KQ971371 EFA09985.1 KQ435794 KOX73893.1 ATLV01010846 ATLV01010847 KE524620 KFB35769.1 KZ288189 PBC34631.1 ADMH02001866 ETN60751.1 KQ434827 KZC07602.1 KQ414713 KOC62970.1 GBYB01003583 JAG73350.1 JTDY01000493 KOB76934.1 GBYB01003584 JAG73351.1 CAQQ02088132 NNAY01000701 OXU26910.1 APGK01050307 KB741169 ENN73324.1 KB629940 ERL83298.1 KB632336 ERL92843.1 GEBQ01028109 JAT11868.1 ACPB03014203 ACPB03014204 ACPB03014205 GL762910 EFZ20475.1 KQ982911 KYQ49328.1 ADTU01023471 KQ976511 KYM82527.1 GL888066 EGI68377.1 KQ980824 KYN12563.1 KQ981280 KYN43461.1 ABLF02039488 GGMR01009004 MBY21623.1 KQ976885 KYN07473.1 KK852543 KDR21673.1 GGMS01007253 MBY76456.1 GL435030 EFN74189.1 GFXV01001671 MBW13476.1 QOIP01000001 RLU27012.1 KK107154 EZA56883.1 GBHO01033665 GBRD01003088 JAG09939.1 JAG62733.1 GBRD01003087 JAG62734.1 GEDC01000231 JAS37067.1 NEVH01006721 PNF37113.1 KFB35770.1 CAQQ02088126 CAQQ02088127 CAQQ02088128 CAQQ02088129 LJIG01000941 KRT85999.1 JRES01001119 KNC25385.1 DS235494 EEB16075.1 ACPB03007015 EAT41377.1 GGMS01003952 MBY73155.1 KK855069 PTY20664.1 GGMR01014986 MBY27605.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000283053
UP000075880
+ More
UP000075886 UP000076407 UP000075903 UP000075882 UP000183832 UP000075883 UP000008820 UP000069940 UP000249989 UP000075902 UP000075840 UP000069272 UP000007266 UP000053105 UP000030765 UP000075881 UP000242457 UP000000673 UP000075900 UP000076502 UP000005203 UP000053825 UP000037510 UP000015102 UP000076408 UP000215335 UP000019118 UP000030742 UP000075885 UP000002358 UP000015103 UP000075809 UP000005205 UP000078540 UP000007755 UP000078492 UP000078541 UP000007819 UP000078542 UP000027135 UP000000311 UP000279307 UP000053097 UP000075920 UP000235965 UP000075901 UP000078200 UP000037069 UP000009046 UP000092443 UP000192223 UP000091820
UP000075886 UP000076407 UP000075903 UP000075882 UP000183832 UP000075883 UP000008820 UP000069940 UP000249989 UP000075902 UP000075840 UP000069272 UP000007266 UP000053105 UP000030765 UP000075881 UP000242457 UP000000673 UP000075900 UP000076502 UP000005203 UP000053825 UP000037510 UP000015102 UP000076408 UP000215335 UP000019118 UP000030742 UP000075885 UP000002358 UP000015103 UP000075809 UP000005205 UP000078540 UP000007755 UP000078492 UP000078541 UP000007819 UP000078542 UP000027135 UP000000311 UP000279307 UP000053097 UP000075920 UP000235965 UP000075901 UP000078200 UP000037069 UP000009046 UP000092443 UP000192223 UP000091820
PRIDE
Pfam
PF00151 Lipase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9J6P5
A0A2H1VRN5
A0A0N1PHU7
A0A194Q079
A0A212FF39
A0A3S2M6M2
+ More
A0A0K8TL87 A0A336K9X1 A0A182JD12 A0A182QP58 A0A2C9H8Q9 A0A1S4H056 A0A182V5D4 A0A182KUQ3 A0A1J1IWQ7 Q8ITU8 A0A182MBT5 A0A1S4FFD6 Q173Z4 A0A182GWJ6 A0A182U9H5 A0A2C9GRV7 A0A182F5H1 D6X109 A0A0M9A1C7 A0A084VCS5 A0A182K8P8 A0A2A3EU98 W5J9S5 A0A182R9Q1 A0A154P6W0 A0A088AGW0 A0A0L7QWJ1 A0A0C9PQR6 A0A0L7LNJ9 A0A0C9QJ60 T1GFZ3 A0A182YFE9 A0A232F8T8 N6SZV3 U4TTN9 A0A182PDD5 K7IRS5 U4UK56 A0A1B6KK83 T1H875 E9IG57 A0A151WNE0 A0A158NQR1 A0A195BEA4 F4WBU9 A0A195DI41 A0A195FSN8 J9K567 A0A2S2NWL8 A0A195D3J4 A0A067RF91 A0A2S2QFF1 E1ZX99 A0A2H8TH74 A0A3L8E398 A0A026WLQ1 A0A182WGZ1 A0A0A9WQ61 A0A0K8TB47 A0A1B6EGK5 A0A2J7R8E4 A0A084VCS6 A0A182PDD4 A0A182TB02 A0A182R9Q0 A0A182YFE8 A0A182MMH0 A0A182QMI5 A0A2C9H8S2 A0A2C9GS68 T1GFZ4 A0A1A9UMK0 A0A0T6BFB5 A0A0L0BZD9 E0VRR9 A0A1A9XZJ2 T1H8M0 A0A1W4WZZ2 Q173Z3 A0A1A9WZW6 A0A2S2Q5X0 A0A2R7WM19 A0A1S4FFC6 A0A2S2PDY1
A0A0K8TL87 A0A336K9X1 A0A182JD12 A0A182QP58 A0A2C9H8Q9 A0A1S4H056 A0A182V5D4 A0A182KUQ3 A0A1J1IWQ7 Q8ITU8 A0A182MBT5 A0A1S4FFD6 Q173Z4 A0A182GWJ6 A0A182U9H5 A0A2C9GRV7 A0A182F5H1 D6X109 A0A0M9A1C7 A0A084VCS5 A0A182K8P8 A0A2A3EU98 W5J9S5 A0A182R9Q1 A0A154P6W0 A0A088AGW0 A0A0L7QWJ1 A0A0C9PQR6 A0A0L7LNJ9 A0A0C9QJ60 T1GFZ3 A0A182YFE9 A0A232F8T8 N6SZV3 U4TTN9 A0A182PDD5 K7IRS5 U4UK56 A0A1B6KK83 T1H875 E9IG57 A0A151WNE0 A0A158NQR1 A0A195BEA4 F4WBU9 A0A195DI41 A0A195FSN8 J9K567 A0A2S2NWL8 A0A195D3J4 A0A067RF91 A0A2S2QFF1 E1ZX99 A0A2H8TH74 A0A3L8E398 A0A026WLQ1 A0A182WGZ1 A0A0A9WQ61 A0A0K8TB47 A0A1B6EGK5 A0A2J7R8E4 A0A084VCS6 A0A182PDD4 A0A182TB02 A0A182R9Q0 A0A182YFE8 A0A182MMH0 A0A182QMI5 A0A2C9H8S2 A0A2C9GS68 T1GFZ4 A0A1A9UMK0 A0A0T6BFB5 A0A0L0BZD9 E0VRR9 A0A1A9XZJ2 T1H8M0 A0A1W4WZZ2 Q173Z3 A0A1A9WZW6 A0A2S2Q5X0 A0A2R7WM19 A0A1S4FFC6 A0A2S2PDY1
PDB
1W52
E-value=7.54747e-17,
Score=211
Ontologies
PANTHER
Topology
Subcellular location
Secreted
Length:
255
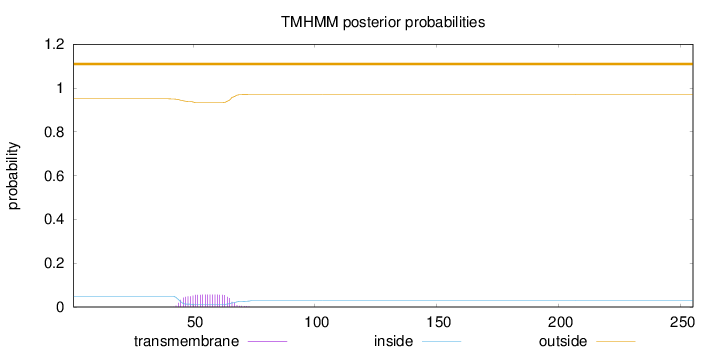
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.18783
Exp number, first 60 AAs:
0.85984
Total prob of N-in:
0.04935
outside
1 - 255
Population Genetic Test Statistics
Pi
19.118167
Theta
21.060418
Tajima's D
-0.464193
CLR
0.288185
CSRT
0.250387480625969
Interpretation
Uncertain
