Gene
KWMTBOMO07456
Pre Gene Modal
BGIBMGA005186
Annotation
dpy-30-like_protein_[Danaus_plexippus]
Full name
Protein dpy-30 homolog
Alternative Name
Dpy-30-like protein
Protein rAIP1
Protein rAIP1
Location in the cell
Nuclear Reliability : 2.57
Sequence
CDS
ATGTCAGAAAGTTCAACACAGATAAATCACAACAAGGAAGAACAATCCACGACGAAAATACCAGATTCGGTAAAGAAAATTATATTAATGGAAAAAGACAATGAAGCCAATGCTAACAGAAAATCCAGAATTGACCTCAATGCTCTACCGACTCGACAGTATTTGGACCAGACAGTAGTACCCATACTGCTGCAAGGTCTATCAGCATTAGCTAAAGAACGTCCTCCGGACCCGATAAATTATTTAGCAGCTTACTTACTTAAAAACAAAACTACTTTCGAACTTGCCAACTCAGGCAATAACAATAATCAACCAACAAACCCACAAACATAA
Protein
MSESSTQINHNKEEQSTTKIPDSVKKIILMEKDNEANANRKSRIDLNALPTRQYLDQTVVPILLQGLSALAKERPPDPINYLAAYLLKNKTTFELANSGNNNNQPTNPQT
Summary
Description
As part of the MLL1/MLL complex, involved in the methylation of histone H3 at 'Lys-4', particularly trimethylation. Histone H3 'Lys-4' methylation represents a specific tag for epigenetic transcriptional activation. May play some role in histone H3 acetylation. In embryonic stem (ES) cells, plays a crucial role in the differentiation potential, particularly along the neural lineage, regulating gene induction and histone H3 'Lys-4' methylation at key developmental loci, including that mediated by retinoic acid. Does not affect ES cell self-renewal. May also play an indirect or direct role in endosomal transport.
As part of the MLL1/MLL complex, involved in the methylation of histone H3 at 'Lys-4', particularly trimethylation. Histone H3 'Lys-4' methylation represents a specific tag for epigenetic transcriptional activation. May play some role in histone H3 acetylation. In a teratocarcinoma cell, plays a crucial role in retinoic acid-induced differentiation along the neural lineage, regulating gene induction and H3 'Lys-4' methylation at key developmental loci. May also play an indirect or direct role in endosomal transport.
As part of the MLL1/MLL complex, involved in the methylation of histone H3 at 'Lys-4', particularly trimethylation. Histone H3 'Lys-4' methylation represents a specific tag for epigenetic transcriptional activation. May play some role in histone H3 acetylation. In embryonic stem cells, may play a crucial role in retinoic acid-induced differentiation along the neural lineage, regulating gene induction and H3 'Lys-4' methylation at key developmental loci. May also play an indirect or direct role in endosomal transport (By similarity).
As part of the MLL1/MLL complex, involved in the methylation of histone H3 at 'Lys-4', particularly trimethylation. Histone H3 'Lys-4' methylation represents a specific tag for epigenetic transcriptional activation. May play some role in histone H3 acetylation. In a teratocarcinoma cell, plays a crucial role in retinoic acid-induced differentiation along the neural lineage, regulating gene induction and H3 'Lys-4' methylation at key developmental loci. May also play an indirect or direct role in endosomal transport.
As part of the MLL1/MLL complex, involved in the methylation of histone H3 at 'Lys-4', particularly trimethylation. Histone H3 'Lys-4' methylation represents a specific tag for epigenetic transcriptional activation. May play some role in histone H3 acetylation. In embryonic stem cells, may play a crucial role in retinoic acid-induced differentiation along the neural lineage, regulating gene induction and H3 'Lys-4' methylation at key developmental loci. May also play an indirect or direct role in endosomal transport (By similarity).
Subunit
Homodimer. Core component of several methyltransferase-containing complexes including MLL1/MLL, MLL2/3 (also named ASCOM complex) and MLL4/WBP7. Each complex is at least composed of ASH2L, RBBP5, WDR5, DPY30, one or more specific histone methyltransferases (KMT2A/MLL1, KMT2D/MLL2, KMT2C/MLL3 and KMT2B/MLL4), and the facultative components MEN1, HCFC1, HCFC2, NCOA6, KDM6A, PAXIP1/PTIP, PAGR1 and alpha- and beta-tubulin PAXIP1/PTIP, PAGR1 and alpha- and beta-tubulin. Interacts with ASH2L. The interaction with ASH2L is direct (By similarity). Interacts with ARFGEF1 (By similarity). Component of the SET1 complex, at least composed of the catalytic subunit (SETD1A or SETD1B), WDR5, WDR82, RBBP5, ASH2L/ASH2, CXXC1/CFP1, HCFC1 and DPY30 (By similarity).
Homodimer. Core component of several methyltransferase-containing complexes including MLL1/MLL, MLL2/3 (also named ASCOM complex) and MLL4/WBP7. Each complex is at least composed of ASH2L, RBBP5, WDR5, DPY30, one or more specific histone methyltransferases (KMT2A/MLL1, KMT2D/MLL2, KMT2C/MLL3 and KMT2B/MLL4), and the facultative components MEN1, HCFC1, HCFC2, NCOA6, KDM6A, PAXIP1/PTIP, PAGR1 and alpha- and beta-tubulin (By similarity). Interacts with ASH2L; the interaction is direct. Interacts with ARFGEF1. Component of the SET1 complex, at least composed of the catalytic subunit (SETD1A or SETD1B), WDR5, WDR82, RBBP5, ASH2L/ASH2, CXXC1/CFP1, HCFC1 and DPY30.
Homodimer. Core component of several methyltransferase-containing complexes including MLL1/MLL, MLL2/3 (also named ASCOM complex) and MLL4/WBP7. Each complex is at least composed of ASH2L, RBBP5, WDR5, DPY30, one or more specific histone methyltransferases (KMT2A/MLL1, KMT2D/MLL2, KMT2C/MLL3 and KMT2B/MLL4), and the facultative components MEN1, HCFC1, HCFC2, NCOA6, KDM6A, PAXIP1/PTIP, PAGR1 and alpha- and beta-tubulin (By similarity). Interacts with ASH2L; the interaction is direct (By similarity). Interacts with ARFGEF1 (By similarity). Component of the SET1 complex, at least composed of the catalytic subunit (SETD1A or SETD1B), WDR5, WDR82, RBBP5, ASH2L/ASH2, CXXC1/CFP1, HCFC1 and DPY30 (By similarity).
Homodimer. Core component of several methyltransferase-containing complexes including MLL1/MLL, MLL2/3 (also named ASCOM complex) and MLL4/WBP7. Each complex is at least composed of ASH2L, RBBP5, WDR5, DPY30, one or more specific histone methyltransferases (KMT2A/MLL1, KMT2D/MLL2, KMT2C/MLL3 and KMT2B/MLL4), and the facultative components MEN1, HCFC1, HCFC2, NCOA6, KDM6A, PAXIP1/PTIP, PAGR1 and alpha- and beta-tubulin (By similarity). Interacts with ASH2L; the interaction is direct (By similarity). Interacts with ARFGEF1. Component of the SET1 complex, at least composed of the catalytic subunit (SETD1A or SETD1B), WDR5, WDR82, RBBP5, ASH2L/ASH2, CXXC1/CFP1, HCFC1 and DPY30 (By similarity).
Homodimer. Core component of several methyltransferase-containing complexes including MLL1/MLL, MLL2/3 (also named ASCOM complex) and MLL4/WBP7. Each complex is at least composed of ASH2L, RBBP5, WDR5, DPY30, one or more specific histone methyltransferases (KMT2A/MLL1, KMT2D/MLL2, KMT2C/MLL3 and KMT2B/MLL4), and the facultative components MEN1, HCFC1, HCFC2, NCOA6, KDM6A, PAXIP1/PTIP, PAGR1 and alpha- and beta-tubulin (By similarity). Interacts with ASH2L; the interaction is direct. Interacts with ARFGEF1. Component of the SET1 complex, at least composed of the catalytic subunit (SETD1A or SETD1B), WDR5, WDR82, RBBP5, ASH2L/ASH2, CXXC1/CFP1, HCFC1 and DPY30.
Homodimer. Core component of several methyltransferase-containing complexes including MLL1/MLL, MLL2/3 (also named ASCOM complex) and MLL4/WBP7. Each complex is at least composed of ASH2L, RBBP5, WDR5, DPY30, one or more specific histone methyltransferases (KMT2A/MLL1, KMT2D/MLL2, KMT2C/MLL3 and KMT2B/MLL4), and the facultative components MEN1, HCFC1, HCFC2, NCOA6, KDM6A, PAXIP1/PTIP, PAGR1 and alpha- and beta-tubulin (By similarity). Interacts with ASH2L; the interaction is direct (By similarity). Interacts with ARFGEF1 (By similarity). Component of the SET1 complex, at least composed of the catalytic subunit (SETD1A or SETD1B), WDR5, WDR82, RBBP5, ASH2L/ASH2, CXXC1/CFP1, HCFC1 and DPY30 (By similarity).
Homodimer. Core component of several methyltransferase-containing complexes including MLL1/MLL, MLL2/3 (also named ASCOM complex) and MLL4/WBP7. Each complex is at least composed of ASH2L, RBBP5, WDR5, DPY30, one or more specific histone methyltransferases (KMT2A/MLL1, KMT2D/MLL2, KMT2C/MLL3 and KMT2B/MLL4), and the facultative components MEN1, HCFC1, HCFC2, NCOA6, KDM6A, PAXIP1/PTIP, PAGR1 and alpha- and beta-tubulin (By similarity). Interacts with ASH2L; the interaction is direct (By similarity). Interacts with ARFGEF1. Component of the SET1 complex, at least composed of the catalytic subunit (SETD1A or SETD1B), WDR5, WDR82, RBBP5, ASH2L/ASH2, CXXC1/CFP1, HCFC1 and DPY30 (By similarity).
Similarity
Belongs to the dpy-30 family.
Keywords
Acetylation
Chromatin regulator
Complete proteome
Golgi apparatus
Isopeptide bond
Nucleus
Phosphoprotein
Reference proteome
Transcription
Transcription regulation
Ubl conjugation
3D-structure
Feature
chain Protein dpy-30 homolog
Uniprot
H9J6P4
A0A212FF19
A0A0N1PJ52
A0A2A4IW39
A0A3S2LFH6
A0A1E1WRM3
+ More
A0A2H1WG58 A0A194PV06 S4PTU4 A0A0L7LNA3 A0A224XPG0 A0A1B6LV27 A0A1B6EXH9 A0A1B6J9X0 R4UM91 K7J696 A0A210Q476 A0A232F7M3 G3V946 Q99LT0 A0A2Y9D8U1 A0A1S3KFV2 G3TMD2 Q640A0 L5KSA4 H0XLH0 Q7TQ15 A0A1U7QJL4 G3HEW0 L8IP41 I3MKV3 S9X4L9 A0A250Y8T2 A0A091E3C4 A0A2K6D6Y8 A0A2K6MEC1 G1TEM5 G1PS50 A0A2K5VRJ2 A0A2I2YG65 A0A2I3GIG7 A0A2K5YRN3 F2Z5F7 A0A2R9BZ79 A0A096NAA2 A0A2K6RH52 A0A2K5LFS2 A0A2K5QQQ1 A0A2K5I343 A0A2K6UQ96 A0A2K6FTU1 S7NSS7 A0A2Y9EGV5 A0A2J8VNI6 A0A341AEA4 A0A2U3V2Z1 A0A340WNB9 A0A384AVL8 F7GJI8 H2QHQ3 A0A2Y9N562 F7IR99 Q9C005 Q2NKU6 Q8K3E7 A0A2Y9HMQ2 A0A2U3W6E4 A0A2U3XK28 A0A3Q7NQ85 F7DY89 U3EBH7 G1LMF2 E2R3B5 M3X6U1 A0A3P4M6F6 M3Y1H3 A0A3Q7V8Z4 A0A2Y9K8A7 A0A384CUF0 A0A3Q7T4C7 U3CSU9 A0A2C9L0V2 A0A212CXE6 A0A1L8G7T8 H0V7L1 A0A1S3EYH7 A0A1S2ZSP3 A0A0P6J5J3 Q6GQK3 A0A091I0N7
A0A2H1WG58 A0A194PV06 S4PTU4 A0A0L7LNA3 A0A224XPG0 A0A1B6LV27 A0A1B6EXH9 A0A1B6J9X0 R4UM91 K7J696 A0A210Q476 A0A232F7M3 G3V946 Q99LT0 A0A2Y9D8U1 A0A1S3KFV2 G3TMD2 Q640A0 L5KSA4 H0XLH0 Q7TQ15 A0A1U7QJL4 G3HEW0 L8IP41 I3MKV3 S9X4L9 A0A250Y8T2 A0A091E3C4 A0A2K6D6Y8 A0A2K6MEC1 G1TEM5 G1PS50 A0A2K5VRJ2 A0A2I2YG65 A0A2I3GIG7 A0A2K5YRN3 F2Z5F7 A0A2R9BZ79 A0A096NAA2 A0A2K6RH52 A0A2K5LFS2 A0A2K5QQQ1 A0A2K5I343 A0A2K6UQ96 A0A2K6FTU1 S7NSS7 A0A2Y9EGV5 A0A2J8VNI6 A0A341AEA4 A0A2U3V2Z1 A0A340WNB9 A0A384AVL8 F7GJI8 H2QHQ3 A0A2Y9N562 F7IR99 Q9C005 Q2NKU6 Q8K3E7 A0A2Y9HMQ2 A0A2U3W6E4 A0A2U3XK28 A0A3Q7NQ85 F7DY89 U3EBH7 G1LMF2 E2R3B5 M3X6U1 A0A3P4M6F6 M3Y1H3 A0A3Q7V8Z4 A0A2Y9K8A7 A0A384CUF0 A0A3Q7T4C7 U3CSU9 A0A2C9L0V2 A0A212CXE6 A0A1L8G7T8 H0V7L1 A0A1S3EYH7 A0A1S2ZSP3 A0A0P6J5J3 Q6GQK3 A0A091I0N7
Pubmed
19121390
22118469
26354079
23622113
26227816
20075255
+ More
28812685 28648823 15057822 15632090 16141072 15489334 21183079 21335234 20431018 23258410 21804562 29704459 22751099 23149746 28087693 21993624 22398555 30723633 22722832 25362486 17431167 25319552 16136131 25243066 14992727 17500065 18838538 19413330 19556245 19651892 19608861 21269460 22814378 24275569 28112733 19481096 19892987 20010809 16341006 17975172 24813606 15562597 27762356
28812685 28648823 15057822 15632090 16141072 15489334 21183079 21335234 20431018 23258410 21804562 29704459 22751099 23149746 28087693 21993624 22398555 30723633 22722832 25362486 17431167 25319552 16136131 25243066 14992727 17500065 18838538 19413330 19556245 19651892 19608861 21269460 22814378 24275569 28112733 19481096 19892987 20010809 16341006 17975172 24813606 15562597 27762356
EMBL
BABH01038151
AGBW02008864
OWR52341.1
KQ460636
KPJ13385.1
NWSH01006480
+ More
PCG63402.1 RSAL01000020 RVE52627.1 GDQN01001603 JAT89451.1 ODYU01008350 SOQ51862.1 KQ459592 KPI96823.1 GAIX01011433 JAA81127.1 JTDY01000493 KOB76932.1 GFTR01002050 JAW14376.1 GEBQ01012455 JAT27522.1 GECZ01027055 JAS42714.1 GECU01011734 JAS95972.1 KC632406 AGM32220.1 NEDP02005067 OWF43548.1 NNAY01000798 OXU26448.1 AC139392 CH473947 EDM02841.1 AK003688 AK011643 AK013073 AK077902 BC002240 AAMC01001126 AAMC01001127 BC082731 CR760705 AAH82731.1 CAJ81411.1 KB030580 ELK14110.1 AAQR03049743 AAQR03049744 AAQR03049745 AY297538 AAP85259.1 JH000321 RAZU01000247 EGW02708.1 RLQ62637.1 JH880854 ELR58340.1 AGTP01024915 AGTP01024916 KB016924 EPY82419.1 GFFW01004761 JAV40027.1 KN121053 KFO37183.1 AAGW02005810 AAPE02028502 AQIA01018625 CABD030013583 ADFV01011574 ADFV01011575 AEMK02000021 DQIR01115934 DQIR01268845 HDA71410.1 AJFE02095359 AJFE02095360 AJFE02095361 AJFE02095362 AJFE02095363 AHZZ02005142 AHZZ02005143 KE164790 EPQ20131.1 ABGA01121802 ABGA01121803 ABGA01121804 ABGA01121805 ABGA01191737 NDHI03003415 PNJ59073.1 PNJ59074.1 JSUE03010519 JSUE03010520 JSUE03010521 JSUE03010522 JU320223 JU473496 JV048468 AFE63979.1 AFH30300.1 AFI38539.1 AACZ04002030 GABC01007852 GABF01010123 GABD01009796 GABE01011611 NBAG03000318 JAA03486.1 JAA12022.1 JAA23304.1 JAA33128.1 PNI41261.1 PNI41262.1 GAMS01009741 GAMR01005581 GAMQ01005389 JAB13395.1 JAB28351.1 JAB36462.1 AF226998 CH471053 BC015970 BC111634 AY129401 GAMP01010843 JAB41912.1 ACTA01129117 ACTA01137117 AAEX03010834 AANG04003404 CYRY02006524 VCW70890.1 AEYP01092831 AEYP01092832 GAMT01007770 JAB04091.1 MKHE01000011 OWK10668.1 CM004474 OCT79968.1 AAKN02023527 GEBF01005773 JAN97859.1 BC072739 CM004475 AAH72739.1 OCT77933.1 KL218009 KFP00973.1
PCG63402.1 RSAL01000020 RVE52627.1 GDQN01001603 JAT89451.1 ODYU01008350 SOQ51862.1 KQ459592 KPI96823.1 GAIX01011433 JAA81127.1 JTDY01000493 KOB76932.1 GFTR01002050 JAW14376.1 GEBQ01012455 JAT27522.1 GECZ01027055 JAS42714.1 GECU01011734 JAS95972.1 KC632406 AGM32220.1 NEDP02005067 OWF43548.1 NNAY01000798 OXU26448.1 AC139392 CH473947 EDM02841.1 AK003688 AK011643 AK013073 AK077902 BC002240 AAMC01001126 AAMC01001127 BC082731 CR760705 AAH82731.1 CAJ81411.1 KB030580 ELK14110.1 AAQR03049743 AAQR03049744 AAQR03049745 AY297538 AAP85259.1 JH000321 RAZU01000247 EGW02708.1 RLQ62637.1 JH880854 ELR58340.1 AGTP01024915 AGTP01024916 KB016924 EPY82419.1 GFFW01004761 JAV40027.1 KN121053 KFO37183.1 AAGW02005810 AAPE02028502 AQIA01018625 CABD030013583 ADFV01011574 ADFV01011575 AEMK02000021 DQIR01115934 DQIR01268845 HDA71410.1 AJFE02095359 AJFE02095360 AJFE02095361 AJFE02095362 AJFE02095363 AHZZ02005142 AHZZ02005143 KE164790 EPQ20131.1 ABGA01121802 ABGA01121803 ABGA01121804 ABGA01121805 ABGA01191737 NDHI03003415 PNJ59073.1 PNJ59074.1 JSUE03010519 JSUE03010520 JSUE03010521 JSUE03010522 JU320223 JU473496 JV048468 AFE63979.1 AFH30300.1 AFI38539.1 AACZ04002030 GABC01007852 GABF01010123 GABD01009796 GABE01011611 NBAG03000318 JAA03486.1 JAA12022.1 JAA23304.1 JAA33128.1 PNI41261.1 PNI41262.1 GAMS01009741 GAMR01005581 GAMQ01005389 JAB13395.1 JAB28351.1 JAB36462.1 AF226998 CH471053 BC015970 BC111634 AY129401 GAMP01010843 JAB41912.1 ACTA01129117 ACTA01137117 AAEX03010834 AANG04003404 CYRY02006524 VCW70890.1 AEYP01092831 AEYP01092832 GAMT01007770 JAB04091.1 MKHE01000011 OWK10668.1 CM004474 OCT79968.1 AAKN02023527 GEBF01005773 JAN97859.1 BC072739 CM004475 AAH72739.1 OCT77933.1 KL218009 KFP00973.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000218220
UP000283053
UP000053268
+ More
UP000037510 UP000002358 UP000242188 UP000215335 UP000002494 UP000000589 UP000248480 UP000085678 UP000007646 UP000008143 UP000010552 UP000005225 UP000189706 UP000001075 UP000273346 UP000005215 UP000028990 UP000233120 UP000233180 UP000001811 UP000001074 UP000233100 UP000001519 UP000001073 UP000233140 UP000008227 UP000240080 UP000028761 UP000233200 UP000233060 UP000233040 UP000233080 UP000233220 UP000233160 UP000248484 UP000001595 UP000252040 UP000245320 UP000265300 UP000261681 UP000006718 UP000002277 UP000248483 UP000008225 UP000005640 UP000009136 UP000248481 UP000245340 UP000245341 UP000286641 UP000002281 UP000008912 UP000002254 UP000011712 UP000000715 UP000286642 UP000248482 UP000261680 UP000291021 UP000286640 UP000076420 UP000186698 UP000005447 UP000081671 UP000079721 UP000054308
UP000037510 UP000002358 UP000242188 UP000215335 UP000002494 UP000000589 UP000248480 UP000085678 UP000007646 UP000008143 UP000010552 UP000005225 UP000189706 UP000001075 UP000273346 UP000005215 UP000028990 UP000233120 UP000233180 UP000001811 UP000001074 UP000233100 UP000001519 UP000001073 UP000233140 UP000008227 UP000240080 UP000028761 UP000233200 UP000233060 UP000233040 UP000233080 UP000233220 UP000233160 UP000248484 UP000001595 UP000252040 UP000245320 UP000265300 UP000261681 UP000006718 UP000002277 UP000248483 UP000008225 UP000005640 UP000009136 UP000248481 UP000245340 UP000245341 UP000286641 UP000002281 UP000008912 UP000002254 UP000011712 UP000000715 UP000286642 UP000248482 UP000261680 UP000291021 UP000286640 UP000076420 UP000186698 UP000005447 UP000081671 UP000079721 UP000054308
Pfam
PF05186 Dpy-30
ProteinModelPortal
H9J6P4
A0A212FF19
A0A0N1PJ52
A0A2A4IW39
A0A3S2LFH6
A0A1E1WRM3
+ More
A0A2H1WG58 A0A194PV06 S4PTU4 A0A0L7LNA3 A0A224XPG0 A0A1B6LV27 A0A1B6EXH9 A0A1B6J9X0 R4UM91 K7J696 A0A210Q476 A0A232F7M3 G3V946 Q99LT0 A0A2Y9D8U1 A0A1S3KFV2 G3TMD2 Q640A0 L5KSA4 H0XLH0 Q7TQ15 A0A1U7QJL4 G3HEW0 L8IP41 I3MKV3 S9X4L9 A0A250Y8T2 A0A091E3C4 A0A2K6D6Y8 A0A2K6MEC1 G1TEM5 G1PS50 A0A2K5VRJ2 A0A2I2YG65 A0A2I3GIG7 A0A2K5YRN3 F2Z5F7 A0A2R9BZ79 A0A096NAA2 A0A2K6RH52 A0A2K5LFS2 A0A2K5QQQ1 A0A2K5I343 A0A2K6UQ96 A0A2K6FTU1 S7NSS7 A0A2Y9EGV5 A0A2J8VNI6 A0A341AEA4 A0A2U3V2Z1 A0A340WNB9 A0A384AVL8 F7GJI8 H2QHQ3 A0A2Y9N562 F7IR99 Q9C005 Q2NKU6 Q8K3E7 A0A2Y9HMQ2 A0A2U3W6E4 A0A2U3XK28 A0A3Q7NQ85 F7DY89 U3EBH7 G1LMF2 E2R3B5 M3X6U1 A0A3P4M6F6 M3Y1H3 A0A3Q7V8Z4 A0A2Y9K8A7 A0A384CUF0 A0A3Q7T4C7 U3CSU9 A0A2C9L0V2 A0A212CXE6 A0A1L8G7T8 H0V7L1 A0A1S3EYH7 A0A1S2ZSP3 A0A0P6J5J3 Q6GQK3 A0A091I0N7
A0A2H1WG58 A0A194PV06 S4PTU4 A0A0L7LNA3 A0A224XPG0 A0A1B6LV27 A0A1B6EXH9 A0A1B6J9X0 R4UM91 K7J696 A0A210Q476 A0A232F7M3 G3V946 Q99LT0 A0A2Y9D8U1 A0A1S3KFV2 G3TMD2 Q640A0 L5KSA4 H0XLH0 Q7TQ15 A0A1U7QJL4 G3HEW0 L8IP41 I3MKV3 S9X4L9 A0A250Y8T2 A0A091E3C4 A0A2K6D6Y8 A0A2K6MEC1 G1TEM5 G1PS50 A0A2K5VRJ2 A0A2I2YG65 A0A2I3GIG7 A0A2K5YRN3 F2Z5F7 A0A2R9BZ79 A0A096NAA2 A0A2K6RH52 A0A2K5LFS2 A0A2K5QQQ1 A0A2K5I343 A0A2K6UQ96 A0A2K6FTU1 S7NSS7 A0A2Y9EGV5 A0A2J8VNI6 A0A341AEA4 A0A2U3V2Z1 A0A340WNB9 A0A384AVL8 F7GJI8 H2QHQ3 A0A2Y9N562 F7IR99 Q9C005 Q2NKU6 Q8K3E7 A0A2Y9HMQ2 A0A2U3W6E4 A0A2U3XK28 A0A3Q7NQ85 F7DY89 U3EBH7 G1LMF2 E2R3B5 M3X6U1 A0A3P4M6F6 M3Y1H3 A0A3Q7V8Z4 A0A2Y9K8A7 A0A384CUF0 A0A3Q7T4C7 U3CSU9 A0A2C9L0V2 A0A212CXE6 A0A1L8G7T8 H0V7L1 A0A1S3EYH7 A0A1S2ZSP3 A0A0P6J5J3 Q6GQK3 A0A091I0N7
PDB
6E2H
E-value=1.71744e-17,
Score=212
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
Golgi apparatus
trans-Golgi network
Golgi apparatus
trans-Golgi network
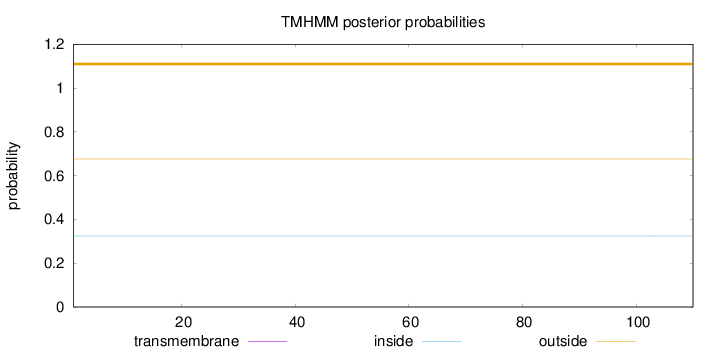
Length:
110
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0034
Exp number, first 60 AAs:
0.00139
Total prob of N-in:
0.32422
outside
1 - 110
Population Genetic Test Statistics
Pi
7.89279
Theta
15.840237
Tajima's D
-1.51159
CLR
8.299891
CSRT
0.0585470726463677
Interpretation
Uncertain
