Pre Gene Modal
BGIBMGA005182
Annotation
PREDICTED:_alpha?alpha-trehalose-phosphate_synthase_[UDP-forming]_isoform_X2_[Bombyx_mori]
Full name
Serine/threonine-protein phosphatase
Location in the cell
Cytoplasmic Reliability : 2.017
Sequence
CDS
ATGTTTCAGATAGTCCCCATTCGCGCGGAACCTAAACTCTTCGACAGCTACTACAATGGATGCTGTAACGGGACATTCTGGCCATTGTTTCATTCGATGCCAGACCGTGCCACTTTCATTGCTGACCACTGGAAGGCATATATCAAGATAAACGAGGAGTTTGCTGAGAAGACAGTCCACGCTCTCCATCTGCTGAACAAACAAAAGAAAGACAAAAATGGAAGTCCACCGATCGTGTGGGTTCACGACTACCATCTGATGTTGGCGGCTAATTGGATACGACAGAGAGCTGAAGAAGATGAGATTAAATGCAAGCTAGCATTCTTTCTGCACATACCGTTTCCACCGTGGGATATTTTTAGACTGTTCCCGTGGTCTGATGAAGTTTTGCAGGGAATTTTGGGTTGTGATATGGTGGGATTCCACATAGAGGATTACTGTTTGAATTTCATTGACTGCTGCCAAAGAAACTTGGGTTGCCGTGTCGATAGAAAAAATCTTCTGGTCGAACTGGGAGATCGCACCATCTGCGTGAGACCGCTACCGATCGGAGTGCCATACGAGAGATTCGTACAGCTGGCCCAAAACGCTAAGCCCGTGTTCTCTACAAGCCAAAAAGTGATATTAGGTGTTGATAGACTGGATTATACCAAAGGATTAGTGCATAGGTTGAAAGCTTTCGAGAGATTACTCGAGAAACACCCGGAGCATATCGAAAAGGTTGTTCTGCTCCAGATCTCGGTGCCCTCTAGGACAGATGTGAAGGAATACCAAGATCTGAAGGAGGAGATGGATCAGCTCGTTGGCAGGATCAATGGTCGATTTACAACGCCCAATTGGTCGCCGATAAGATATATTTACGGCTGCGTTGGTCAAGATGAGTTAGCGGCGTTTTATCGCGATGCTGCTGTGGCACTTGTAACTCCATTAAGAGACGGCATGAACCTGGTCGCGAAAGAGTTTGTGGCTTGTCAGATCAATAAGCCTCCTGGAGTATTAATAGTGTCGCCATTTGCTGGAGCAGGAGAAATGATGCACGAGGCTCTTATATGCAATCCTTATGAGCTCGACGATGCGGCCGAAGTCATTCATAGAGCCTTAATAATGCCAGAGGATGAACGGACAGTCCGCATGAACCACCTCAGGCGGAGGGAGCAGTTGAACGACGTGGACAGCTGGATGAAGGCGTTCCTCAAAGCGATGGACTCTCTGGAGGAAGAGGCCGACGACCTTGGAGCCACTACCATGCAACCAGTCACTATTGACGATTTCGATGAATATTTATCCAAGTACATCGGGTACACACAGAAGCTAGCCCTGCTGTTGGACTACGACGGTACCTTGGCCCCGATCGCGCCCCACCCGGACCTCGCCACGCTGCCTCTCGAGACCAAACACACCCTGCAGAGGCTCTCCAACATGTCCGATGTGTACATAGCCATCATATCGGGGAGGAACGTGGACAACGTCAAGCAAATGGTCGGTATCGAAGGCATCACGTATGCGGGTAACCACGGTCTGGAGATCTTACATCCTGACGGCAGCAAGTTTGTCCATCCAATGCCGATGGAGCTGCAAGACAAGGTCGTTGACCTGCTCAAATCACTGCAGGAGCAGGTATGCAAAGACGGAGCTTGGGTTGAGAACAAAGGAGCGTTGCTAACGTTCCACTACCGCGAGACTCCCGTGCCGAAGCGGGCCGCGCTGGCCGAGCAGGCCCGCAAGCTCATCACGGCCTCGGGCTTCACCCCGGCGCCGGCGCACTGCGCGCTGGAGGCGCGGCCCCCGGTGCAATGGGACAAAGGACGAGCTTCGATCTACATCTTGAGGACAGCATTCGGTTTGGACTGGAGCGAAAGGATCAGGATCATATACGCCGGTGACGACAACACCGACGAAGACGCTATGCTGGCTTTAAAAGGTATGGCTGCCACCTTCCGCATCGTATCATCGAACATCATAAAAACGTCGGCAGAACGACGTTTGTCCTCCACCGACTCTGTACTGGCGATGCTGAAGTGGGTCGAACGCCACTTCTCGAGACGCAAGCCCAGAGCGAATTCACTAACGTACAAGAACGCCAGGAAGGCCAGGGATACAATACAGATGCATATGTCATATCAAATGCCTTCGAAGACATCGCCCAGACACTCTCCCCCACATACTCCGGACAAAGGCTCAAGCGGCTCGGACTCGAGCTAA
Protein
MFQIVPIRAEPKLFDSYYNGCCNGTFWPLFHSMPDRATFIADHWKAYIKINEEFAEKTVHALHLLNKQKKDKNGSPPIVWVHDYHLMLAANWIRQRAEEDEIKCKLAFFLHIPFPPWDIFRLFPWSDEVLQGILGCDMVGFHIEDYCLNFIDCCQRNLGCRVDRKNLLVELGDRTICVRPLPIGVPYERFVQLAQNAKPVFSTSQKVILGVDRLDYTKGLVHRLKAFERLLEKHPEHIEKVVLLQISVPSRTDVKEYQDLKEEMDQLVGRINGRFTTPNWSPIRYIYGCVGQDELAAFYRDAAVALVTPLRDGMNLVAKEFVACQINKPPGVLIVSPFAGAGEMMHEALICNPYELDDAAEVIHRALIMPEDERTVRMNHLRRREQLNDVDSWMKAFLKAMDSLEEEADDLGATTMQPVTIDDFDEYLSKYIGYTQKLALLLDYDGTLAPIAPHPDLATLPLETKHTLQRLSNMSDVYIAIISGRNVDNVKQMVGIEGITYAGNHGLEILHPDGSKFVHPMPMELQDKVVDLLKSLQEQVCKDGAWVENKGALLTFHYRETPVPKRAALAEQARKLITASGFTPAPAHCALEARPPVQWDKGRASIYILRTAFGLDWSERIRIIYAGDDNTDEDAMLALKGMAATFRIVSSNIIKTSAERRLSSTDSVLAMLKWVERHFSRRKPRANSLTYKNARKARDTIQMHMSYQMPSKTSPRHSPPHTPDKGSSGSDSS
Summary
Catalytic Activity
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
Similarity
Belongs to the PPP phosphatase family.
Uniprot
H9J6P0
A0A2H1WFJ9
D2KHI8
A0A2A4ISC9
A0A194PU34
B5TD29
+ More
A4GHG0 A0A2D0WL60 A0A0N1I975 A0A3G2SGU2 A0A2D3E2A0 A0A1J0M552 A0A2D3E238 S4NTI3 A0A212FF13 A0A1W6MPR2 J7G8S5 A0A310SSI4 V9IA83 K7IWD1 A0A0L7RFM2 A0A2A3EC11 A0A088A3B7 E2BI26 A0A151JTW4 A0A195EDF4 F4WQ53 A0A0C9RFD4 A0A158NX63 E2ANA9 A0A0C9QKI7 A0A026WD64 A0A151WYT2 A0A151I569 A0A0J7L2L4 A0A151I6N7 A0A1B0CHY7 A0A084W4K2 E0W3W6 A0A1Y0AWT0 A0A232F8Y6 A0A182J7Z1 A0A182K209 A0A2M4BE41 A0A2M4BDZ4 A0A2M4BDZ6 A0A1Q3FSB7 U5ESG9 D2A2U8 A0A139WIW2 A0A1D8I2M3 A0A1P8VK81 A0A182NFF1 V5G5Y4 Q176D3 A0A2M3ZFV5 A0A2M4A4G8 A0A182F160 W5JRT9 A0A182UM34 A0A182TWM4 A0A182XL08 A0A182L142 Q7Q455 A0A182HUE6 A0A2M3YZX5 A0A182GNU7 A0A182RX95 A0A182Y3K7 A0A182PF95 A0A182QJ39 A0A1L8E1Q7 A0A182W6U1 A0A182MG12 C8CCZ9 A0A023EVW9 A0A1B6M5T0 A0A1B0D3L9 A8D372 A0A1B6DMS5 B0WQL4 C8CGD1 A0A0L0C9U2 A0A0T6BDN0 I3UII1 A0A0K8TMP5 W8BEA7 U4UEP4 W8AI07 A0A034W4R7 W8B4D0 A0A034W337 J9JJR9 A0A0K8W1I3 J9WJ78 A0A0A1XDM0 A0A1B1PFM0 J9K2L1 T1PD62 A0A1A9WUV1
A4GHG0 A0A2D0WL60 A0A0N1I975 A0A3G2SGU2 A0A2D3E2A0 A0A1J0M552 A0A2D3E238 S4NTI3 A0A212FF13 A0A1W6MPR2 J7G8S5 A0A310SSI4 V9IA83 K7IWD1 A0A0L7RFM2 A0A2A3EC11 A0A088A3B7 E2BI26 A0A151JTW4 A0A195EDF4 F4WQ53 A0A0C9RFD4 A0A158NX63 E2ANA9 A0A0C9QKI7 A0A026WD64 A0A151WYT2 A0A151I569 A0A0J7L2L4 A0A151I6N7 A0A1B0CHY7 A0A084W4K2 E0W3W6 A0A1Y0AWT0 A0A232F8Y6 A0A182J7Z1 A0A182K209 A0A2M4BE41 A0A2M4BDZ4 A0A2M4BDZ6 A0A1Q3FSB7 U5ESG9 D2A2U8 A0A139WIW2 A0A1D8I2M3 A0A1P8VK81 A0A182NFF1 V5G5Y4 Q176D3 A0A2M3ZFV5 A0A2M4A4G8 A0A182F160 W5JRT9 A0A182UM34 A0A182TWM4 A0A182XL08 A0A182L142 Q7Q455 A0A182HUE6 A0A2M3YZX5 A0A182GNU7 A0A182RX95 A0A182Y3K7 A0A182PF95 A0A182QJ39 A0A1L8E1Q7 A0A182W6U1 A0A182MG12 C8CCZ9 A0A023EVW9 A0A1B6M5T0 A0A1B0D3L9 A8D372 A0A1B6DMS5 B0WQL4 C8CGD1 A0A0L0C9U2 A0A0T6BDN0 I3UII1 A0A0K8TMP5 W8BEA7 U4UEP4 W8AI07 A0A034W4R7 W8B4D0 A0A034W337 J9JJR9 A0A0K8W1I3 J9WJ78 A0A0A1XDM0 A0A1B1PFM0 J9K2L1 T1PD62 A0A1A9WUV1
EC Number
3.1.3.16
Pubmed
19121390
26354079
19004876
18492231
20193689
23622113
+ More
22118469 22762304 20075255 20798317 21719571 21347285 24508170 30249741 24438588 20566863 28341416 28648823 18362917 19820115 17510324 20920257 23761445 20966253 12364791 14747013 17210077 26483478 25244985 24945155 26108605 26369729 24495485 23537049 25348373 22972362 25830018 25315136
22118469 22762304 20075255 20798317 21719571 21347285 24508170 30249741 24438588 20566863 28341416 28648823 18362917 19820115 17510324 20920257 23761445 20966253 12364791 14747013 17210077 26483478 25244985 24945155 26108605 26369729 24495485 23537049 25348373 22972362 25830018 25315136
EMBL
BABH01038155
BABH01038156
ODYU01008350
SOQ51859.1
GU211889
ADA63844.1
+ More
NWSH01008242 PCG62641.1 KQ459592 KPI96827.1 EU878265 ACH88521.1 EF051258 EU647215 ABM66814.2 ACD01424.1 KU977454 ARD05072.1 KQ460636 KPJ13381.1 MG787167 AYO46920.1 KY565574 ATU31386.1 KX013356 APD15513.1 KY565578 ATU31390.1 GAIX01012206 JAA80354.1 AGBW02008864 OWR52345.1 KX037427 ARN79552.1 JX025053 AFP67548.1 KQ760495 OAD60116.1 JR037900 JR037902 AEY58022.1 AEY58023.1 KQ414606 KOC69531.1 KZ288293 PBC29014.1 GL448433 EFN84652.1 KQ981799 KYN35667.1 KQ979074 KYN22879.1 GL888262 EGI63676.1 GBYB01015274 JAG85041.1 ADTU01003009 GL441177 EFN65057.1 GBYB01015278 JAG85045.1 KK107260 QOIP01000001 EZA54015.1 RLU27505.1 KQ982651 KYQ52855.1 KQ976434 KYM87221.1 LBMM01001097 KMQ96931.1 KQ978464 KYM93770.1 AJWK01012907 AJWK01012908 AJWK01012909 ATLV01020343 KE525299 KFB45146.1 DS235883 EEB20322.1 KY921835 ART29424.1 NNAY01000634 OXU27286.1 GGFJ01002122 MBW51263.1 GGFJ01002124 MBW51265.1 GGFJ01002123 MBW51264.1 GFDL01004722 JAV30323.1 GANO01003249 JAB56622.1 KQ971338 EFA02222.2 KYB27852.1 KU756283 AOT99586.1 KU578006 APZ77037.1 GALX01002996 JAB65470.1 CH477390 EAT41968.1 GGFM01006638 MBW27389.1 GGFK01002291 MBW35612.1 ADMH02000541 ETN66003.1 AAAB01008964 EAA12459.4 APCN01002455 GGFM01001045 MBW21796.1 JXUM01077203 KQ562993 KXJ74691.1 AXCN02000817 GFDF01001421 JAV12663.1 AXCM01003220 GQ389790 ACV32626.1 GAPW01000230 JAC13368.1 GEBQ01008697 JAT31280.1 AJVK01002975 EU131894 ABV44614.1 GEDC01010330 JAS26968.1 DS232041 EDS32889.1 GQ397450 ACV20871.1 JRES01000714 KNC29021.1 LJIG01001488 KRT85452.1 JQ743627 AFK64819.1 GDAI01001994 JAI15609.1 GAMC01018431 JAB88124.1 KB632144 ERL89081.1 GAMC01018430 JAB88125.1 GAKP01009822 JAC49130.1 GAMC01018429 JAB88126.1 GAKP01009823 JAC49129.1 ABLF02021486 GDHF01007322 JAI44992.1 JX462664 AFS17320.1 GBXI01005242 JAD09050.1 KU379749 ANT46147.1 ABLF02006957 ABLF02006958 KA646727 AFP61356.1
NWSH01008242 PCG62641.1 KQ459592 KPI96827.1 EU878265 ACH88521.1 EF051258 EU647215 ABM66814.2 ACD01424.1 KU977454 ARD05072.1 KQ460636 KPJ13381.1 MG787167 AYO46920.1 KY565574 ATU31386.1 KX013356 APD15513.1 KY565578 ATU31390.1 GAIX01012206 JAA80354.1 AGBW02008864 OWR52345.1 KX037427 ARN79552.1 JX025053 AFP67548.1 KQ760495 OAD60116.1 JR037900 JR037902 AEY58022.1 AEY58023.1 KQ414606 KOC69531.1 KZ288293 PBC29014.1 GL448433 EFN84652.1 KQ981799 KYN35667.1 KQ979074 KYN22879.1 GL888262 EGI63676.1 GBYB01015274 JAG85041.1 ADTU01003009 GL441177 EFN65057.1 GBYB01015278 JAG85045.1 KK107260 QOIP01000001 EZA54015.1 RLU27505.1 KQ982651 KYQ52855.1 KQ976434 KYM87221.1 LBMM01001097 KMQ96931.1 KQ978464 KYM93770.1 AJWK01012907 AJWK01012908 AJWK01012909 ATLV01020343 KE525299 KFB45146.1 DS235883 EEB20322.1 KY921835 ART29424.1 NNAY01000634 OXU27286.1 GGFJ01002122 MBW51263.1 GGFJ01002124 MBW51265.1 GGFJ01002123 MBW51264.1 GFDL01004722 JAV30323.1 GANO01003249 JAB56622.1 KQ971338 EFA02222.2 KYB27852.1 KU756283 AOT99586.1 KU578006 APZ77037.1 GALX01002996 JAB65470.1 CH477390 EAT41968.1 GGFM01006638 MBW27389.1 GGFK01002291 MBW35612.1 ADMH02000541 ETN66003.1 AAAB01008964 EAA12459.4 APCN01002455 GGFM01001045 MBW21796.1 JXUM01077203 KQ562993 KXJ74691.1 AXCN02000817 GFDF01001421 JAV12663.1 AXCM01003220 GQ389790 ACV32626.1 GAPW01000230 JAC13368.1 GEBQ01008697 JAT31280.1 AJVK01002975 EU131894 ABV44614.1 GEDC01010330 JAS26968.1 DS232041 EDS32889.1 GQ397450 ACV20871.1 JRES01000714 KNC29021.1 LJIG01001488 KRT85452.1 JQ743627 AFK64819.1 GDAI01001994 JAI15609.1 GAMC01018431 JAB88124.1 KB632144 ERL89081.1 GAMC01018430 JAB88125.1 GAKP01009822 JAC49130.1 GAMC01018429 JAB88126.1 GAKP01009823 JAC49129.1 ABLF02021486 GDHF01007322 JAI44992.1 JX462664 AFS17320.1 GBXI01005242 JAD09050.1 KU379749 ANT46147.1 ABLF02006957 ABLF02006958 KA646727 AFP61356.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000002358
+ More
UP000053825 UP000242457 UP000005203 UP000008237 UP000078541 UP000078492 UP000007755 UP000005205 UP000000311 UP000053097 UP000279307 UP000075809 UP000078540 UP000036403 UP000078542 UP000092461 UP000030765 UP000009046 UP000215335 UP000075880 UP000075881 UP000007266 UP000075884 UP000008820 UP000069272 UP000000673 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000069940 UP000249989 UP000075900 UP000076408 UP000075885 UP000075886 UP000075920 UP000075883 UP000092462 UP000002320 UP000037069 UP000030742 UP000007819 UP000095301 UP000091820
UP000053825 UP000242457 UP000005203 UP000008237 UP000078541 UP000078492 UP000007755 UP000005205 UP000000311 UP000053097 UP000279307 UP000075809 UP000078540 UP000036403 UP000078542 UP000092461 UP000030765 UP000009046 UP000215335 UP000075880 UP000075881 UP000007266 UP000075884 UP000008820 UP000069272 UP000000673 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000069940 UP000249989 UP000075900 UP000076408 UP000075885 UP000075886 UP000075920 UP000075883 UP000092462 UP000002320 UP000037069 UP000030742 UP000007819 UP000095301 UP000091820
PRIDE
Pfam
Interpro
IPR023214
HAD_sf
+ More
IPR001830 Glyco_trans_20
IPR036412 HAD-like_sf
IPR003337 Trehalose_PPase
IPR006379 HAD-SF_hydro_IIB
IPR005835 NTP_transferase_dom
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR029044 Nucleotide-diphossugar_trans
IPR035985 Ubiquitin-activating_enz
IPR000594 ThiF_NAD_FAD-bd
IPR029752 D-isomer_DH_CS1
IPR001451 Hexapep
IPR004843 Calcineurin-like_PHP_ApaH
IPR006186 Ser/Thr-sp_prot-phosphatase
IPR029052 Metallo-depent_PP-like
IPR001830 Glyco_trans_20
IPR036412 HAD-like_sf
IPR003337 Trehalose_PPase
IPR006379 HAD-SF_hydro_IIB
IPR005835 NTP_transferase_dom
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR029044 Nucleotide-diphossugar_trans
IPR035985 Ubiquitin-activating_enz
IPR000594 ThiF_NAD_FAD-bd
IPR029752 D-isomer_DH_CS1
IPR001451 Hexapep
IPR004843 Calcineurin-like_PHP_ApaH
IPR006186 Ser/Thr-sp_prot-phosphatase
IPR029052 Metallo-depent_PP-like
Gene 3D
CDD
ProteinModelPortal
H9J6P0
A0A2H1WFJ9
D2KHI8
A0A2A4ISC9
A0A194PU34
B5TD29
+ More
A4GHG0 A0A2D0WL60 A0A0N1I975 A0A3G2SGU2 A0A2D3E2A0 A0A1J0M552 A0A2D3E238 S4NTI3 A0A212FF13 A0A1W6MPR2 J7G8S5 A0A310SSI4 V9IA83 K7IWD1 A0A0L7RFM2 A0A2A3EC11 A0A088A3B7 E2BI26 A0A151JTW4 A0A195EDF4 F4WQ53 A0A0C9RFD4 A0A158NX63 E2ANA9 A0A0C9QKI7 A0A026WD64 A0A151WYT2 A0A151I569 A0A0J7L2L4 A0A151I6N7 A0A1B0CHY7 A0A084W4K2 E0W3W6 A0A1Y0AWT0 A0A232F8Y6 A0A182J7Z1 A0A182K209 A0A2M4BE41 A0A2M4BDZ4 A0A2M4BDZ6 A0A1Q3FSB7 U5ESG9 D2A2U8 A0A139WIW2 A0A1D8I2M3 A0A1P8VK81 A0A182NFF1 V5G5Y4 Q176D3 A0A2M3ZFV5 A0A2M4A4G8 A0A182F160 W5JRT9 A0A182UM34 A0A182TWM4 A0A182XL08 A0A182L142 Q7Q455 A0A182HUE6 A0A2M3YZX5 A0A182GNU7 A0A182RX95 A0A182Y3K7 A0A182PF95 A0A182QJ39 A0A1L8E1Q7 A0A182W6U1 A0A182MG12 C8CCZ9 A0A023EVW9 A0A1B6M5T0 A0A1B0D3L9 A8D372 A0A1B6DMS5 B0WQL4 C8CGD1 A0A0L0C9U2 A0A0T6BDN0 I3UII1 A0A0K8TMP5 W8BEA7 U4UEP4 W8AI07 A0A034W4R7 W8B4D0 A0A034W337 J9JJR9 A0A0K8W1I3 J9WJ78 A0A0A1XDM0 A0A1B1PFM0 J9K2L1 T1PD62 A0A1A9WUV1
A4GHG0 A0A2D0WL60 A0A0N1I975 A0A3G2SGU2 A0A2D3E2A0 A0A1J0M552 A0A2D3E238 S4NTI3 A0A212FF13 A0A1W6MPR2 J7G8S5 A0A310SSI4 V9IA83 K7IWD1 A0A0L7RFM2 A0A2A3EC11 A0A088A3B7 E2BI26 A0A151JTW4 A0A195EDF4 F4WQ53 A0A0C9RFD4 A0A158NX63 E2ANA9 A0A0C9QKI7 A0A026WD64 A0A151WYT2 A0A151I569 A0A0J7L2L4 A0A151I6N7 A0A1B0CHY7 A0A084W4K2 E0W3W6 A0A1Y0AWT0 A0A232F8Y6 A0A182J7Z1 A0A182K209 A0A2M4BE41 A0A2M4BDZ4 A0A2M4BDZ6 A0A1Q3FSB7 U5ESG9 D2A2U8 A0A139WIW2 A0A1D8I2M3 A0A1P8VK81 A0A182NFF1 V5G5Y4 Q176D3 A0A2M3ZFV5 A0A2M4A4G8 A0A182F160 W5JRT9 A0A182UM34 A0A182TWM4 A0A182XL08 A0A182L142 Q7Q455 A0A182HUE6 A0A2M3YZX5 A0A182GNU7 A0A182RX95 A0A182Y3K7 A0A182PF95 A0A182QJ39 A0A1L8E1Q7 A0A182W6U1 A0A182MG12 C8CCZ9 A0A023EVW9 A0A1B6M5T0 A0A1B0D3L9 A8D372 A0A1B6DMS5 B0WQL4 C8CGD1 A0A0L0C9U2 A0A0T6BDN0 I3UII1 A0A0K8TMP5 W8BEA7 U4UEP4 W8AI07 A0A034W4R7 W8B4D0 A0A034W337 J9JJR9 A0A0K8W1I3 J9WJ78 A0A0A1XDM0 A0A1B1PFM0 J9K2L1 T1PD62 A0A1A9WUV1
PDB
5HUV
E-value=4.48131e-81,
Score=770
Ontologies
PATHWAY
GO
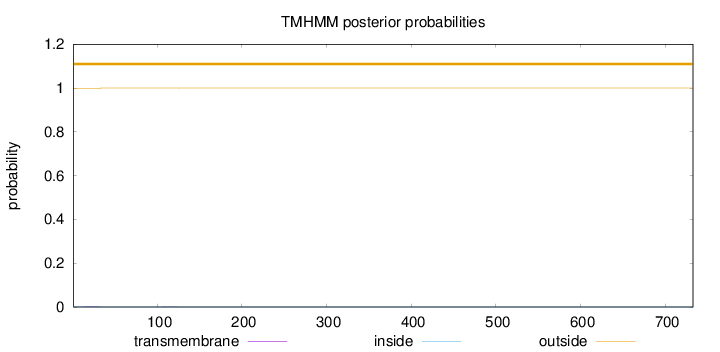
Topology
Length:
733
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0434400000000001
Exp number, first 60 AAs:
0.03888
Total prob of N-in:
0.00234
outside
1 - 733
Population Genetic Test Statistics
Pi
20.262643
Theta
18.657715
Tajima's D
-1.707167
CLR
1.07757
CSRT
0.0357982100894955
Interpretation
Uncertain
