Gene
KWMTBOMO07448
Annotation
PREDICTED:_zinc_finger_BED_domain-containing_protein_1-like_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.763 Nuclear Reliability : 1.878
Sequence
CDS
ATGGATGATGGAGTACTTCAGTTTCAACCTGTTCCAAGTTGCAGTGGCAGTCAAAGATCCGGTGCACCGTTACCAAAAACCGTAAATGAAACCATTGATTCAGACTTAGACGAAAACGTTACACCTTTAAAACGCCAAAGAACAATACAGGCCGCTTTTAAAGATATTTCCTCTTACTCAGAATCTGGAGCGAAGACAAGGAAACTGAACAACTGTTTGTTATTCATGATTTGCAAAGACCATCAGCCATTCTCAATCGTTGAAAACGAAGGCTTCAAAACTCTAATGAAAACAGTTGCTCCTCAATATAAGCTCCCGAGTAGAACAACTCTAAGACGCTGGCTGGACGATAAATATGAAGTAATTTCAGAAACAATTAAGAAAATGTTGTCCTCTATAGAAGATATAACCCTAACAACAGATATTTGGTCGGATACCTTGAATATGAAAAGTTTTTTGGGCATCACAGCACATTTCGGAATAGACATTGAACTATATTCAGTCACCTTAGGTGTGTACGAATTGGATGAAAGACATACATCAGATTATCTTTCTGAAATTCTCTTAAAGGTATGTCAAGACTGGAAAATCGATCAAGAGAATGTAACAGCTGTTGTTACTGACAACGCAGCCAATATAGTTAAGGCGATCGAATTAGCGTTTGGTAGAAGGAAACACATCCCATGTTTTGCCCATACTCTGAATTTGGTGGCTGAAGGCACAATGATATGTATTGAGTGGAGAAACATTGTTTCTAAAGTAAAATCAATAGTGACTTGGTTTAAACAGAGTTGCATTGCTAGTGATGAATTACGTAAAGCTACTGCTGCGGCTACGGAGTCTTCTGCTGGAGCAGCTGAGGTAAAATTAATACAAAGCGTGGATACTCGCTGGAACAGTACATATTATATGTTACAAAGATTTCTAGAACTCCGTAGTGTCATAAATGATATTCTCTTCCGCCACCCTAGAGCACCTGCAATGCTGAGTGCATCGGAAATATCTACTGTATCGTCTGTCATCATGGTATTGCGACCCTTAGAAGCGGCTACAAAAGAAATTTCAGGTGATAAATACTGCACAAGTAGTAAAATTATTCCCTTAGTTCGTTGCATGCTCTCTAAAATAACTTCAGCTGTTATAGAAGATCCCGTAGCGAAAGAGGTGCAGAAACTTGCTATTAATGAAATAAATAAAAGAATGGGTTCAATAGAACATGTGAATGCTCTGGCTATCGCAAGTATTTTAGATCCACGGTTTAAAAGAATGCATTTCAATGATCCGATAGCTTGTTCGAATGCTGTTACGAAAATTAAAGATTTAATGAAGAAGAATTTGCATAGCAATGAAGAACCTGAGTCCGATTCTGACAAGTCCGATAAAAATGAAGAGGCCTTTTCATTATGGACTGATCACCATAAGTTAGTGCATCGTAACTGGAAAATTAACAAATCTGAAGACGTTTTGTCTGACGAACTTTCCGTTTATCTCCGAACTCCTGTTGGAAGACTTAAAGAAAATCCGTTAGAAATTTGGAGAGATTACAAAATACAATTCCCGAAGCTGTACAAAATAGCTTTTAAGTATTTAACAATAGTTGGCACGTCAGTCCCTTCAGAAAGACTTTTTTCACAGGCAGGGCTAGTTATGACTGAACAAAGAAACCGGCTGAAGGGGAAGAGATTAAGCAAACTTTTGTTTTTACATAGCATTGACAAAAAATATTGGAATATGTAA
Protein
MDDGVLQFQPVPSCSGSQRSGAPLPKTVNETIDSDLDENVTPLKRQRTIQAAFKDISSYSESGAKTRKLNNCLLFMICKDHQPFSIVENEGFKTLMKTVAPQYKLPSRTTLRRWLDDKYEVISETIKKMLSSIEDITLTTDIWSDTLNMKSFLGITAHFGIDIELYSVTLGVYELDERHTSDYLSEILLKVCQDWKIDQENVTAVVTDNAANIVKAIELAFGRRKHIPCFAHTLNLVAEGTMICIEWRNIVSKVKSIVTWFKQSCIASDELRKATAAATESSAGAAEVKLIQSVDTRWNSTYYMLQRFLELRSVINDILFRHPRAPAMLSASEISTVSSVIMVLRPLEAATKEISGDKYCTSSKIIPLVRCMLSKITSAVIEDPVAKEVQKLAINEINKRMGSIEHVNALAIASILDPRFKRMHFNDPIACSNAVTKIKDLMKKNLHSNEEPESDSDKSDKNEEAFSLWTDHHKLVHRNWKINKSEDVLSDELSVYLRTPVGRLKENPLEIWRDYKIQFPKLYKIAFKYLTIVGTSVPSERLFSQAGLVMTEQRNRLKGKRLSKLLFLHSIDKKYWNM
Summary
Uniprot
J9LDY8
A0A1W4WLZ1
A0A1E1WN08
J9M7K2
A0A151J3A2
J9M2M5
+ More
E2AN53 A0A151IJ50 A0A151IHU3 J9KF88 J9M5N4 E2C2R7 A0A151J8J8 A0A151ICL6 A0A195DX32 A0A151IVN1 A0A151K1W2 A0A0J7K4Y0 A0A2S2QDH1 A0A2S2R3S9 J9KNP5 J9L2Y9 A0A3L8DVM9 J9LSA5 A0A026WKR9 A0A026X0S0 A0A151IK53 J9LFC3 J9LEV9 J9LVM0 J9KS03 A0A151WQ96 A0A224XKI7 T1JXM7 E2A4Y2 A0A2S2QCJ4 Q8T648 A0A1X7UCW0 A0A158P559 A0A1X7TGY6 A0A2S2PB02 A0A1X7UFA9 X1WTG7 T1K5U2 J9L6T0 A0A026VWT7 A0A0J7KSL2 J9LS52 A0A0T6AT01 G3PR68 J9M5D4 A0A1X7TEQ5 A0A3P8RVY2 J9LRC3 A0A1A8AM29 A0A3B1JJI0 J9LZV6 J9KQT7 A0A1A8UJB2 J9KBT0 A0A3B4XPZ3 J9L784 A0A1X7SRE6 A0A3B3BJP2 A0A0L7LFD6 A0A232EFR6 A0A3B5QTD9 A0A2A4JMY2 A0A3Q3GI05 A0A1A8FQM2 A0A2A4JLR2 K7JPR8 A0A3P9CU72 H9JJ46 A0A3P8RG16 A0A0A1XSV1 A0A3B3WPP4 A0A3P8PZW8 A0A146ZTV1 A0A3B1J7G0
E2AN53 A0A151IJ50 A0A151IHU3 J9KF88 J9M5N4 E2C2R7 A0A151J8J8 A0A151ICL6 A0A195DX32 A0A151IVN1 A0A151K1W2 A0A0J7K4Y0 A0A2S2QDH1 A0A2S2R3S9 J9KNP5 J9L2Y9 A0A3L8DVM9 J9LSA5 A0A026WKR9 A0A026X0S0 A0A151IK53 J9LFC3 J9LEV9 J9LVM0 J9KS03 A0A151WQ96 A0A224XKI7 T1JXM7 E2A4Y2 A0A2S2QCJ4 Q8T648 A0A1X7UCW0 A0A158P559 A0A1X7TGY6 A0A2S2PB02 A0A1X7UFA9 X1WTG7 T1K5U2 J9L6T0 A0A026VWT7 A0A0J7KSL2 J9LS52 A0A0T6AT01 G3PR68 J9M5D4 A0A1X7TEQ5 A0A3P8RVY2 J9LRC3 A0A1A8AM29 A0A3B1JJI0 J9LZV6 J9KQT7 A0A1A8UJB2 J9KBT0 A0A3B4XPZ3 J9L784 A0A1X7SRE6 A0A3B3BJP2 A0A0L7LFD6 A0A232EFR6 A0A3B5QTD9 A0A2A4JMY2 A0A3Q3GI05 A0A1A8FQM2 A0A2A4JLR2 K7JPR8 A0A3P9CU72 H9JJ46 A0A3P8RG16 A0A0A1XSV1 A0A3B3WPP4 A0A3P8PZW8 A0A146ZTV1 A0A3B1J7G0
Pubmed
EMBL
ABLF02010709
GDQN01002674
JAT88380.1
ABLF02008875
KQ980290
KYN16919.1
+ More
ABLF02006015 GL441080 EFN65135.1 KQ977395 KYN02984.1 KQ977557 KYN02009.1 ABLF02030582 ABLF02001734 GL452203 EFN77752.1 KQ979507 KYN21183.1 KQ978037 KYM97822.1 KQ980204 KYN17277.1 KQ980913 KYN11465.1 LKEX01009590 KYN50115.1 LBMM01014067 KMQ85349.1 GGMS01006357 MBY75560.1 GGMS01015207 MBY84410.1 ABLF02023260 ABLF02009488 QOIP01000003 RLU24471.1 ABLF02017802 KK107161 EZA56605.1 KK107050 EZA61658.1 KQ977255 KYN04655.1 ABLF02011645 ABLF02044591 ABLF02050984 ABLF02055777 ABLF02018406 ABLF02006624 ABLF02006626 ABLF02030547 KQ982846 KYQ49981.1 GFTR01007466 JAW08960.1 CAEY01000832 GL436768 EFN71507.1 GGMS01006230 MBY75433.1 AF486809 AAL93203.1 CAEY01000199 GGMR01013943 MBY26562.1 ABLF02011646 CAEY01001590 ABLF02017539 ABLF02017541 ABLF02047606 ABLF02059536 KK107725 EZA47986.1 LBMM01003701 KMQ93224.1 ABLF02021118 LJIG01022898 KRT78170.1 ABLF02025797 ABLF02021063 ABLF02051044 HADY01017248 SBP55733.1 ABLF02030460 ABLF02030462 ABLF02030463 ABLF02018503 ABLF02031891 HAEJ01007957 SBS48414.1 ABLF02014501 ABLF02031655 JTDY01001348 KOB74120.1 NNAY01004948 OXU17200.1 NWSH01001073 PCG72760.1 HAEB01013902 SBQ60429.1 PCG72759.1 AAZX01004655 BABH01040014 GBXI01000181 JAD14111.1 GCES01016583 JAR69740.1
ABLF02006015 GL441080 EFN65135.1 KQ977395 KYN02984.1 KQ977557 KYN02009.1 ABLF02030582 ABLF02001734 GL452203 EFN77752.1 KQ979507 KYN21183.1 KQ978037 KYM97822.1 KQ980204 KYN17277.1 KQ980913 KYN11465.1 LKEX01009590 KYN50115.1 LBMM01014067 KMQ85349.1 GGMS01006357 MBY75560.1 GGMS01015207 MBY84410.1 ABLF02023260 ABLF02009488 QOIP01000003 RLU24471.1 ABLF02017802 KK107161 EZA56605.1 KK107050 EZA61658.1 KQ977255 KYN04655.1 ABLF02011645 ABLF02044591 ABLF02050984 ABLF02055777 ABLF02018406 ABLF02006624 ABLF02006626 ABLF02030547 KQ982846 KYQ49981.1 GFTR01007466 JAW08960.1 CAEY01000832 GL436768 EFN71507.1 GGMS01006230 MBY75433.1 AF486809 AAL93203.1 CAEY01000199 GGMR01013943 MBY26562.1 ABLF02011646 CAEY01001590 ABLF02017539 ABLF02017541 ABLF02047606 ABLF02059536 KK107725 EZA47986.1 LBMM01003701 KMQ93224.1 ABLF02021118 LJIG01022898 KRT78170.1 ABLF02025797 ABLF02021063 ABLF02051044 HADY01017248 SBP55733.1 ABLF02030460 ABLF02030462 ABLF02030463 ABLF02018503 ABLF02031891 HAEJ01007957 SBS48414.1 ABLF02014501 ABLF02031655 JTDY01001348 KOB74120.1 NNAY01004948 OXU17200.1 NWSH01001073 PCG72760.1 HAEB01013902 SBQ60429.1 PCG72759.1 AAZX01004655 BABH01040014 GBXI01000181 JAD14111.1 GCES01016583 JAR69740.1
Proteomes
UP000007819
UP000192223
UP000078492
UP000000311
UP000078542
UP000008237
+ More
UP000036403 UP000279307 UP000053097 UP000075809 UP000015104 UP000007879 UP000007635 UP000265080 UP000018467 UP000261360 UP000261560 UP000037510 UP000215335 UP000002852 UP000218220 UP000261660 UP000002358 UP000265160 UP000005204 UP000265100 UP000261480
UP000036403 UP000279307 UP000053097 UP000075809 UP000015104 UP000007879 UP000007635 UP000265080 UP000018467 UP000261360 UP000261560 UP000037510 UP000215335 UP000002852 UP000218220 UP000261660 UP000002358 UP000265160 UP000005204 UP000265100 UP000261480
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
J9LDY8
A0A1W4WLZ1
A0A1E1WN08
J9M7K2
A0A151J3A2
J9M2M5
+ More
E2AN53 A0A151IJ50 A0A151IHU3 J9KF88 J9M5N4 E2C2R7 A0A151J8J8 A0A151ICL6 A0A195DX32 A0A151IVN1 A0A151K1W2 A0A0J7K4Y0 A0A2S2QDH1 A0A2S2R3S9 J9KNP5 J9L2Y9 A0A3L8DVM9 J9LSA5 A0A026WKR9 A0A026X0S0 A0A151IK53 J9LFC3 J9LEV9 J9LVM0 J9KS03 A0A151WQ96 A0A224XKI7 T1JXM7 E2A4Y2 A0A2S2QCJ4 Q8T648 A0A1X7UCW0 A0A158P559 A0A1X7TGY6 A0A2S2PB02 A0A1X7UFA9 X1WTG7 T1K5U2 J9L6T0 A0A026VWT7 A0A0J7KSL2 J9LS52 A0A0T6AT01 G3PR68 J9M5D4 A0A1X7TEQ5 A0A3P8RVY2 J9LRC3 A0A1A8AM29 A0A3B1JJI0 J9LZV6 J9KQT7 A0A1A8UJB2 J9KBT0 A0A3B4XPZ3 J9L784 A0A1X7SRE6 A0A3B3BJP2 A0A0L7LFD6 A0A232EFR6 A0A3B5QTD9 A0A2A4JMY2 A0A3Q3GI05 A0A1A8FQM2 A0A2A4JLR2 K7JPR8 A0A3P9CU72 H9JJ46 A0A3P8RG16 A0A0A1XSV1 A0A3B3WPP4 A0A3P8PZW8 A0A146ZTV1 A0A3B1J7G0
E2AN53 A0A151IJ50 A0A151IHU3 J9KF88 J9M5N4 E2C2R7 A0A151J8J8 A0A151ICL6 A0A195DX32 A0A151IVN1 A0A151K1W2 A0A0J7K4Y0 A0A2S2QDH1 A0A2S2R3S9 J9KNP5 J9L2Y9 A0A3L8DVM9 J9LSA5 A0A026WKR9 A0A026X0S0 A0A151IK53 J9LFC3 J9LEV9 J9LVM0 J9KS03 A0A151WQ96 A0A224XKI7 T1JXM7 E2A4Y2 A0A2S2QCJ4 Q8T648 A0A1X7UCW0 A0A158P559 A0A1X7TGY6 A0A2S2PB02 A0A1X7UFA9 X1WTG7 T1K5U2 J9L6T0 A0A026VWT7 A0A0J7KSL2 J9LS52 A0A0T6AT01 G3PR68 J9M5D4 A0A1X7TEQ5 A0A3P8RVY2 J9LRC3 A0A1A8AM29 A0A3B1JJI0 J9LZV6 J9KQT7 A0A1A8UJB2 J9KBT0 A0A3B4XPZ3 J9L784 A0A1X7SRE6 A0A3B3BJP2 A0A0L7LFD6 A0A232EFR6 A0A3B5QTD9 A0A2A4JMY2 A0A3Q3GI05 A0A1A8FQM2 A0A2A4JLR2 K7JPR8 A0A3P9CU72 H9JJ46 A0A3P8RG16 A0A0A1XSV1 A0A3B3WPP4 A0A3P8PZW8 A0A146ZTV1 A0A3B1J7G0
PDB
6DX0
E-value=1.04626e-22,
Score=265
Ontologies
GO
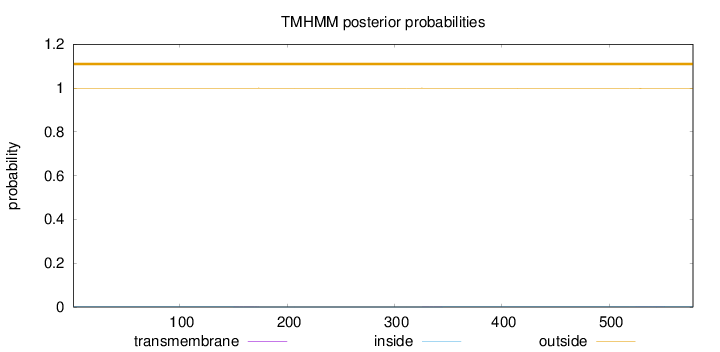
Topology
Length:
578
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03762
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00068
outside
1 - 578
Population Genetic Test Statistics
Pi
1.109648
Theta
2.733908
Tajima's D
-1.332848
CLR
1.743343
CSRT
0.0735463226838658
Interpretation
Uncertain
