Gene
KWMTBOMO07447
Annotation
PREDICTED:_androgen-dependent_TFPI-regulating_protein-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.436
Sequence
CDS
ATGGCAGATATAGTATTTTGGACGACGATGATAACCAACCCGGAGATGATGGCTCCACCGAAACTATTCGACTACCTCCCGTACTGGTGCCAGCACTCGCTGCACACCGTGTCGGCGGTCGTTGTAATCGGAGACTTGATACTGACTCCAAGGAGGAGGCCAAGGAATCTTGCACCGCGTTTCACGATTATGACAGCTTTTTATTTGTTTTATCTCATTGTGCTATACATCAATTATCTGAACGGACAAATAGTGTATAACGCTATCGATAGTCTACCAGGCTCCAAAATAAGAACTATATCCTTGGCCGTTTACACCGGCTACGTGGTTTTCTTTTATTTACAGTGGATGTTAATAGATCTTGTGTGGAGATAG
Protein
MADIVFWTTMITNPEMMAPPKLFDYLPYWCQHSLHTVSAVVVIGDLILTPRRRPRNLAPRFTIMTAFYLFYLIVLYINYLNGQIVYNAIDSLPGSKIRTISLAVYTGYVVFFYLQWMLIDLVWR
Summary
Uniprot
A0A2H1VJG0
A0A194PV09
A0A212FPH7
A0A2A4JWJ9
A0A2H1VK66
A0A194R135
+ More
A0A194PHE5 A0A2A4JBG6 A0A212EPN7 A0A026WE41 E2AN14 A0A195BSL9 A0A3L8E1G6 R0KXE9 U3IZK8 B3MUK1 A0A195DHZ3 B4ICU7 B4Q617 A0A158NQM2 Q9VPM9 A0A195D3P2 A0A2A4ISD3 A0A195EQU0 E2B9U0 A0A3S2LKN5 A0A151XB65 A0A2P2I1V6 A0A194PTG9 A0A0L7R8P0 B4P2B9 A0A1W4UQ09 B3N7N1 B4LUC3 A0A1S3DUL7 A0A1S3DWF7 D0QWD5 F4WKG3 A0A088APY4 B4KGW2 A0A2A3EIC0 V9IL99 A0A1D5NVB8 Q29NW7 A0A3B0JI19 A0A1U7RIT8 K7J3G6 L9L9E7 A0A1L8E3B5 A0A232ETD4 A0A1E1W437 A0A154NY70 A0A1L8E347 A0A1L8E342 A0A183BSH0 A0A1B6CQT4 F6WZQ9 A0A1B6DLQ4 A0A3B3ZYD9 A0A3Q0K6J2 A0A091SWB0 A0A316ZCX1 A0A0K8TF02 H9J137 A0A2W1BWR4 A0A0A9YE34 A0A146LI07 A0A2R7VPQ2 A0A0A9YCL9 A0A0A9YFL9 A0A0A0AAZ9 A0A267DBZ3 A0A182EQD9 A0A0T6BDD8 A0A0M3QT33 A0A2B4R9A3 V8NR98 A0A091JL79 A0A1L8ELP2 A0A091NSI2 A0A091USB7 A0A093RSH8
A0A194PHE5 A0A2A4JBG6 A0A212EPN7 A0A026WE41 E2AN14 A0A195BSL9 A0A3L8E1G6 R0KXE9 U3IZK8 B3MUK1 A0A195DHZ3 B4ICU7 B4Q617 A0A158NQM2 Q9VPM9 A0A195D3P2 A0A2A4ISD3 A0A195EQU0 E2B9U0 A0A3S2LKN5 A0A151XB65 A0A2P2I1V6 A0A194PTG9 A0A0L7R8P0 B4P2B9 A0A1W4UQ09 B3N7N1 B4LUC3 A0A1S3DUL7 A0A1S3DWF7 D0QWD5 F4WKG3 A0A088APY4 B4KGW2 A0A2A3EIC0 V9IL99 A0A1D5NVB8 Q29NW7 A0A3B0JI19 A0A1U7RIT8 K7J3G6 L9L9E7 A0A1L8E3B5 A0A232ETD4 A0A1E1W437 A0A154NY70 A0A1L8E347 A0A1L8E342 A0A183BSH0 A0A1B6CQT4 F6WZQ9 A0A1B6DLQ4 A0A3B3ZYD9 A0A3Q0K6J2 A0A091SWB0 A0A316ZCX1 A0A0K8TF02 H9J137 A0A2W1BWR4 A0A0A9YE34 A0A146LI07 A0A2R7VPQ2 A0A0A9YCL9 A0A0A9YFL9 A0A0A0AAZ9 A0A267DBZ3 A0A182EQD9 A0A0T6BDD8 A0A0M3QT33 A0A2B4R9A3 V8NR98 A0A091JL79 A0A1L8ELP2 A0A091NSI2 A0A091USB7 A0A093RSH8
Pubmed
26354079
22118469
24508170
20798317
30249741
23749191
+ More
17994087 22936249 21347285 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 19859648 21719571 15592404 15632085 20075255 23385571 28648823 18464734 25463417 12203911 29771364 19121390 28756777 25401762 26823975 22919073 24297900 27762356
17994087 22936249 21347285 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 19859648 21719571 15592404 15632085 20075255 23385571 28648823 18464734 25463417 12203911 29771364 19121390 28756777 25401762 26823975 22919073 24297900 27762356
EMBL
ODYU01002885
SOQ40931.1
KQ459592
KPI96828.1
AGBW02002272
OWR55646.1
+ More
NWSH01000443 PCG76397.1 ODYU01003002 SOQ41213.1 KQ460883 KPJ11224.1 KQ459603 KPI92732.1 NWSH01002009 PCG69447.1 AGBW02013442 OWR43427.1 KK107260 EZA54188.1 GL441034 EFN65172.1 KQ976419 KYM89277.1 QOIP01000001 RLU26466.1 KB743575 EOA97963.1 ADON01102522 CH902624 EDV33530.1 KQ980824 KYN12515.1 CH480829 EDW45373.1 CM000361 CM002910 EDX03201.1 KMY87281.1 ADTU01023407 AE014134 AY089240 AAF51517.1 AAL89978.1 AHN54060.1 KQ976885 KYN07518.1 NWSH01008210 PCG62651.1 KQ982021 KYN30578.1 GL446605 EFN87527.1 RSAL01000077 RVE48747.1 KQ982335 KYQ57538.1 IACF01002224 LAB67886.1 KQ459593 KPI96716.1 KQ414632 KOC67136.1 CM000157 EDW87115.1 CH954177 EDV57207.1 CH940649 EDW64109.1 FJ821025 ACN94639.1 GL888199 EGI65301.1 CH933807 EDW11162.1 KZ288232 PBC31543.1 JR051229 AEY61457.1 AADN05000024 CH379058 EAL34526.2 OUUW01000006 SPP81987.1 KB320453 ELW71735.1 GFDF01000938 JAV13146.1 NNAY01002290 OXU21611.1 GDQN01009311 JAT81743.1 KQ434782 KZC04567.1 GFDF01000937 JAV13147.1 GFDF01000936 JAV13148.1 GEDC01021491 JAS15807.1 GEDC01010689 JAS26609.1 KK488158 KFQ62628.1 KZ819292 PWN98093.1 GBRD01001659 JAG64162.1 BABH01031450 KZ149912 PZC78064.1 GBHO01013703 JAG29901.1 GDHC01014258 GDHC01010806 JAQ04371.1 JAQ07823.1 KK854013 PTY09339.1 GBHO01013700 JAG29904.1 GBHO01013701 JAG29903.1 KL871168 KGL91087.1 NIVC01005060 PAA46092.1 UYRW01005859 VDM94077.1 LJIG01002005 KRT84935.1 CP012523 ALC38213.1 LSMT01000817 PFX14221.1 AZIM01002352 ETE64213.1 KK502094 KFP20505.1 CM004483 OCT60241.1 KL392338 KFP92077.1 KL410062 KFQ93621.1 KL411822 KFW73774.1
NWSH01000443 PCG76397.1 ODYU01003002 SOQ41213.1 KQ460883 KPJ11224.1 KQ459603 KPI92732.1 NWSH01002009 PCG69447.1 AGBW02013442 OWR43427.1 KK107260 EZA54188.1 GL441034 EFN65172.1 KQ976419 KYM89277.1 QOIP01000001 RLU26466.1 KB743575 EOA97963.1 ADON01102522 CH902624 EDV33530.1 KQ980824 KYN12515.1 CH480829 EDW45373.1 CM000361 CM002910 EDX03201.1 KMY87281.1 ADTU01023407 AE014134 AY089240 AAF51517.1 AAL89978.1 AHN54060.1 KQ976885 KYN07518.1 NWSH01008210 PCG62651.1 KQ982021 KYN30578.1 GL446605 EFN87527.1 RSAL01000077 RVE48747.1 KQ982335 KYQ57538.1 IACF01002224 LAB67886.1 KQ459593 KPI96716.1 KQ414632 KOC67136.1 CM000157 EDW87115.1 CH954177 EDV57207.1 CH940649 EDW64109.1 FJ821025 ACN94639.1 GL888199 EGI65301.1 CH933807 EDW11162.1 KZ288232 PBC31543.1 JR051229 AEY61457.1 AADN05000024 CH379058 EAL34526.2 OUUW01000006 SPP81987.1 KB320453 ELW71735.1 GFDF01000938 JAV13146.1 NNAY01002290 OXU21611.1 GDQN01009311 JAT81743.1 KQ434782 KZC04567.1 GFDF01000937 JAV13147.1 GFDF01000936 JAV13148.1 GEDC01021491 JAS15807.1 GEDC01010689 JAS26609.1 KK488158 KFQ62628.1 KZ819292 PWN98093.1 GBRD01001659 JAG64162.1 BABH01031450 KZ149912 PZC78064.1 GBHO01013703 JAG29901.1 GDHC01014258 GDHC01010806 JAQ04371.1 JAQ07823.1 KK854013 PTY09339.1 GBHO01013700 JAG29904.1 GBHO01013701 JAG29903.1 KL871168 KGL91087.1 NIVC01005060 PAA46092.1 UYRW01005859 VDM94077.1 LJIG01002005 KRT84935.1 CP012523 ALC38213.1 LSMT01000817 PFX14221.1 AZIM01002352 ETE64213.1 KK502094 KFP20505.1 CM004483 OCT60241.1 KL392338 KFP92077.1 KL410062 KFQ93621.1 KL411822 KFW73774.1
Proteomes
UP000053268
UP000007151
UP000218220
UP000053240
UP000053097
UP000000311
+ More
UP000078540 UP000279307 UP000016666 UP000007801 UP000078492 UP000001292 UP000000304 UP000005205 UP000000803 UP000078542 UP000078541 UP000008237 UP000283053 UP000075809 UP000053825 UP000002282 UP000192221 UP000008711 UP000008792 UP000079169 UP000007755 UP000005203 UP000009192 UP000242457 UP000000539 UP000001819 UP000268350 UP000189705 UP000002358 UP000011518 UP000215335 UP000076502 UP000050741 UP000002279 UP000261520 UP000245946 UP000005204 UP000053858 UP000215902 UP000077448 UP000271087 UP000092553 UP000225706 UP000053119 UP000186698 UP000053283
UP000078540 UP000279307 UP000016666 UP000007801 UP000078492 UP000001292 UP000000304 UP000005205 UP000000803 UP000078542 UP000078541 UP000008237 UP000283053 UP000075809 UP000053825 UP000002282 UP000192221 UP000008711 UP000008792 UP000079169 UP000007755 UP000005203 UP000009192 UP000242457 UP000000539 UP000001819 UP000268350 UP000189705 UP000002358 UP000011518 UP000215335 UP000076502 UP000050741 UP000002279 UP000261520 UP000245946 UP000005204 UP000053858 UP000215902 UP000077448 UP000271087 UP000092553 UP000225706 UP000053119 UP000186698 UP000053283
Pfam
PF04750 Far-17a_AIG1
Interpro
IPR006838
Far-17a_AIG1
ProteinModelPortal
A0A2H1VJG0
A0A194PV09
A0A212FPH7
A0A2A4JWJ9
A0A2H1VK66
A0A194R135
+ More
A0A194PHE5 A0A2A4JBG6 A0A212EPN7 A0A026WE41 E2AN14 A0A195BSL9 A0A3L8E1G6 R0KXE9 U3IZK8 B3MUK1 A0A195DHZ3 B4ICU7 B4Q617 A0A158NQM2 Q9VPM9 A0A195D3P2 A0A2A4ISD3 A0A195EQU0 E2B9U0 A0A3S2LKN5 A0A151XB65 A0A2P2I1V6 A0A194PTG9 A0A0L7R8P0 B4P2B9 A0A1W4UQ09 B3N7N1 B4LUC3 A0A1S3DUL7 A0A1S3DWF7 D0QWD5 F4WKG3 A0A088APY4 B4KGW2 A0A2A3EIC0 V9IL99 A0A1D5NVB8 Q29NW7 A0A3B0JI19 A0A1U7RIT8 K7J3G6 L9L9E7 A0A1L8E3B5 A0A232ETD4 A0A1E1W437 A0A154NY70 A0A1L8E347 A0A1L8E342 A0A183BSH0 A0A1B6CQT4 F6WZQ9 A0A1B6DLQ4 A0A3B3ZYD9 A0A3Q0K6J2 A0A091SWB0 A0A316ZCX1 A0A0K8TF02 H9J137 A0A2W1BWR4 A0A0A9YE34 A0A146LI07 A0A2R7VPQ2 A0A0A9YCL9 A0A0A9YFL9 A0A0A0AAZ9 A0A267DBZ3 A0A182EQD9 A0A0T6BDD8 A0A0M3QT33 A0A2B4R9A3 V8NR98 A0A091JL79 A0A1L8ELP2 A0A091NSI2 A0A091USB7 A0A093RSH8
A0A194PHE5 A0A2A4JBG6 A0A212EPN7 A0A026WE41 E2AN14 A0A195BSL9 A0A3L8E1G6 R0KXE9 U3IZK8 B3MUK1 A0A195DHZ3 B4ICU7 B4Q617 A0A158NQM2 Q9VPM9 A0A195D3P2 A0A2A4ISD3 A0A195EQU0 E2B9U0 A0A3S2LKN5 A0A151XB65 A0A2P2I1V6 A0A194PTG9 A0A0L7R8P0 B4P2B9 A0A1W4UQ09 B3N7N1 B4LUC3 A0A1S3DUL7 A0A1S3DWF7 D0QWD5 F4WKG3 A0A088APY4 B4KGW2 A0A2A3EIC0 V9IL99 A0A1D5NVB8 Q29NW7 A0A3B0JI19 A0A1U7RIT8 K7J3G6 L9L9E7 A0A1L8E3B5 A0A232ETD4 A0A1E1W437 A0A154NY70 A0A1L8E347 A0A1L8E342 A0A183BSH0 A0A1B6CQT4 F6WZQ9 A0A1B6DLQ4 A0A3B3ZYD9 A0A3Q0K6J2 A0A091SWB0 A0A316ZCX1 A0A0K8TF02 H9J137 A0A2W1BWR4 A0A0A9YE34 A0A146LI07 A0A2R7VPQ2 A0A0A9YCL9 A0A0A9YFL9 A0A0A0AAZ9 A0A267DBZ3 A0A182EQD9 A0A0T6BDD8 A0A0M3QT33 A0A2B4R9A3 V8NR98 A0A091JL79 A0A1L8ELP2 A0A091NSI2 A0A091USB7 A0A093RSH8
Ontologies
GO
PANTHER
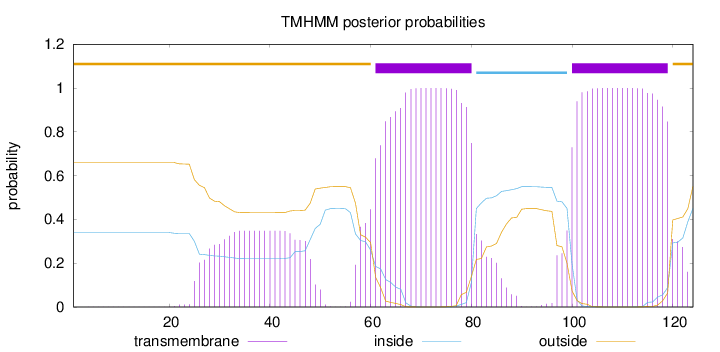
Topology
Length:
124
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
50.2538
Exp number, first 60 AAs:
8.94856
Total prob of N-in:
0.34024
outside
1 - 60
TMhelix
61 - 80
inside
81 - 99
TMhelix
100 - 119
outside
120 - 124
Population Genetic Test Statistics
Pi
6.772357
Theta
4.596128
Tajima's D
-0.660535
CLR
1.43441
CSRT
0.209389530523474
Interpretation
Uncertain
