Pre Gene Modal
BGIBMGA005226
Annotation
ATP-binding_cassette_transporter_subfamily_G_member_Bm3_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.877
Sequence
CDS
ATGAGCACGCGTGCGTCCCGCCTCTCGGGTGGACAGAAGAAGAGGCTCTCCATCGCTCTGGAGCTGATCAACAATCCCCTCGTTATGTTCCTCGACGAACCAACAACTGGTCTGGACAGCTCCTCGTGTAGTCAAGTAGTCAAGCTCTGCAGGGATCTGGCGCAGCAGGGTCGTACCATAGTCTGTACGGTACATCAACCCTCAGCGTCACTGTTTGCCCTATTCGATCAAGTATACGTTTTAGCTGCAGGAAATTGCCTCTACCAGGGCACCACAAAGAACTTAGTACCATACTTGGAGGGGGCCGGAGTACCGTGCCCTATATACCACAATCCAGCTGATTACATTATCGAATTGGCTTGCGGAGAATACGGTGAAGACAAGATCCAAATCCTAACGACAAAAGCTGAAAACGGACGAGCACTTTCCTGGTTCGACGATCCTGACTCCATGCCCACTATGGAAGTCTTAAGAAAGCAGTGTCCTCTTAGCGTCTTAAAGGAGCAAGACCCTAGTACTGGCAACGAAGAAACTCCTAGATCCAATCAAAGAAAAGTACTTATTAAAAGAGGATTCTTAAAAGCAAAACGCGACTCCACAATGACCCATCTCCGGCTAATAGTGAACGTGTTAGTGGGAGTTATGTTGGGGAGTTTATTTATCAAGGCCGGTAACGAAGGCTCCAGAGTCTTGGAGAACTACAATCTGCTGTTTGCGATACTAATGCACCACATGATGTCGCCAATGATGCTTACCATACTCACTTTTCCGTCGGAAATGTCCATATTATCAAAGGAGCACTTCAATAGATGGTACTCGTTGGGTTCCTATTACATCTCTATTACAATTGTCGATTTACCTATTACGATAGTATCTTGTGCGGTGTTCAGTGCGATAGTGTACACTATGTCTGGTCAGCCGATGGCGATCTCTCGGTTCGGGATGTTCTTCGCGATCTCCCTATACACGGTACTAGTGGCGCAGAGTTTCGGCCTCATGATCGGAGCCGTCTTCAACGTTGTTAACGGTACTTTCCTTGGCCCTACTTTGTCGGTACCGATGATGATGTTCGCGGGTTTCGGAGTGACTCTACGCGACCTCCCGCCGTACTTGTACTGGGGGTCATACGTCTCGTACTTGCGGTACGGATTGGAAGGATTCGTGGGCGCCATATACGGGTTGGAAAGACCGAAACTGGACTGCAATGAAGCACCTATATGCCTTTATAAATACCCAAGACAGTTCCTCTCTGAAGTAGCTATGTCTCCGGACAGATTCTGGTGGGACGTTCTAGCGCTCACGATCTTCGTCTTCGTGTTAAGGATTGCAGCTTTCTTCCTCCTAAAATGGAAGTTAAACGCTGTCCGATAA
Protein
MSTRASRLSGGQKKRLSIALELINNPLVMFLDEPTTGLDSSSCSQVVKLCRDLAQQGRTIVCTVHQPSASLFALFDQVYVLAAGNCLYQGTTKNLVPYLEGAGVPCPIYHNPADYIIELACGEYGEDKIQILTTKAENGRALSWFDDPDSMPTMEVLRKQCPLSVLKEQDPSTGNEETPRSNQRKVLIKRGFLKAKRDSTMTHLRLIVNVLVGVMLGSLFIKAGNEGSRVLENYNLLFAILMHHMMSPMMLTILTFPSEMSILSKEHFNRWYSLGSYYISITIVDLPITIVSCAVFSAIVYTMSGQPMAISRFGMFFAISLYTVLVAQSFGLMIGAVFNVVNGTFLGPTLSVPMMMFAGFGVTLRDLPPYLYWGSYVSYLRYGLEGFVGAIYGLERPKLDCNEAPICLYKYPRQFLSEVAMSPDRFWWDVLALTIFVFVLRIAAFFLLKWKLNAVR
Summary
Uniprot
H9J6T4
F2YDQ6
S4PWH7
A0A194PVN9
A0A0N0PCC8
A0A2A4JX93
+ More
A0A2A4JWK0 A0A2H1VKG2 A0A212FPJ2 E0W387 A0A0A1X7A6 A0A0A1X2C7 K7ILU8 A0A034WT57 F4WUW8 A0A1W4XHH8 A0A195AUJ8 A0A151JZL1 A0A158P0V0 A0A0K8V0X6 A0A0C9R177 A0A026WGT2 A0A0C9PL33 A0A232F1V9 A0A151WWP2 T1PB46 A0A0L0CGW6 D2A232 A0A182G655 A0A182G5I4 E9IZD3 Q16Y84 B3MPB8 A0A1S4FK15 A0A182RGB5 A0A182K839 A0A182P7F0 A0A084VPQ7 A0A1I8P8P9 A0A182MVP0 A0A067QV98 W5J3P6 A0A182YG04 A0A182F592 A0A2M4A571 A0A182XFM9 A0A182TXZ8 A0A182UP98 A0A182L5H1 Q7PFM9 A0A182IE85 A0A1Q3FYJ3 B3N8L1 A0A2M4BH59 V5GHI9 B4MX30 A0A2M4A5F4 A0A154NYW5 A0A182JJD9 Q29NF1 Q960D7 A0A2M3Z0C2 A0A182VUV0 A0A2M3Z0Q0 A0A1W4UUD9 A0A0J9QZL2 A0A1Y1KDU5 B4Q8A5 B4NYQ0 Q9VL61 A0A3B0JU51 U4UUH1 B4JCG2 A0A1L8E4I7 B4M9F9 U5EUI7 A0A0M5J908 A0A195CZ41 A0A336M1M3 A0A1A9WHI6 A0A1A9XMQ4 A0A0U9HYG1 B4KH30 A0A1A9VE61 A0A1B0ABB5 A0A1B0G0H7 A0A0L7QX80 A0A2J7RCU0 A0A1B6KVQ4 A0A1B0C1D6 A0A1B0GJ02 A0A1B6EU87 E2A2I4 A0A161ANB1 R4WKD2 A0A0A9YMK9 A0A0A9WFV8 A0A1J1HT67 B4HWD5 V5G0H5
A0A2A4JWK0 A0A2H1VKG2 A0A212FPJ2 E0W387 A0A0A1X7A6 A0A0A1X2C7 K7ILU8 A0A034WT57 F4WUW8 A0A1W4XHH8 A0A195AUJ8 A0A151JZL1 A0A158P0V0 A0A0K8V0X6 A0A0C9R177 A0A026WGT2 A0A0C9PL33 A0A232F1V9 A0A151WWP2 T1PB46 A0A0L0CGW6 D2A232 A0A182G655 A0A182G5I4 E9IZD3 Q16Y84 B3MPB8 A0A1S4FK15 A0A182RGB5 A0A182K839 A0A182P7F0 A0A084VPQ7 A0A1I8P8P9 A0A182MVP0 A0A067QV98 W5J3P6 A0A182YG04 A0A182F592 A0A2M4A571 A0A182XFM9 A0A182TXZ8 A0A182UP98 A0A182L5H1 Q7PFM9 A0A182IE85 A0A1Q3FYJ3 B3N8L1 A0A2M4BH59 V5GHI9 B4MX30 A0A2M4A5F4 A0A154NYW5 A0A182JJD9 Q29NF1 Q960D7 A0A2M3Z0C2 A0A182VUV0 A0A2M3Z0Q0 A0A1W4UUD9 A0A0J9QZL2 A0A1Y1KDU5 B4Q8A5 B4NYQ0 Q9VL61 A0A3B0JU51 U4UUH1 B4JCG2 A0A1L8E4I7 B4M9F9 U5EUI7 A0A0M5J908 A0A195CZ41 A0A336M1M3 A0A1A9WHI6 A0A1A9XMQ4 A0A0U9HYG1 B4KH30 A0A1A9VE61 A0A1B0ABB5 A0A1B0G0H7 A0A0L7QX80 A0A2J7RCU0 A0A1B6KVQ4 A0A1B0C1D6 A0A1B0GJ02 A0A1B6EU87 E2A2I4 A0A161ANB1 R4WKD2 A0A0A9YMK9 A0A0A9WFV8 A0A1J1HT67 B4HWD5 V5G0H5
Pubmed
19121390
23622113
26354079
22118469
20566863
25830018
+ More
20075255 25348373 29121518 21719571 21347285 24508170 30249741 28648823 25315136 26108605 18362917 19820115 26483478 21282665 17510324 17994087 24438588 24845553 20920257 23761445 25244985 20966253 12364791 14747013 17210077 15632085 23185243 22936249 28004739 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23537049 18057021 20798317 23691247 25401762 26823975
20075255 25348373 29121518 21719571 21347285 24508170 30249741 28648823 25315136 26108605 18362917 19820115 26483478 21282665 17510324 17994087 24438588 24845553 20920257 23761445 25244985 20966253 12364791 14747013 17210077 15632085 23185243 22936249 28004739 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23537049 18057021 20798317 23691247 25401762 26823975
EMBL
BABH01037214
BABH01037215
BABH01037216
BABH01037217
BABH01037218
HQ896032
+ More
ADZ56944.1 GAIX01006768 JAA85792.1 KQ459592 KPI96829.1 KQ460636 KPJ13379.1 NWSH01000443 PCG76396.1 PCG76395.1 ODYU01002885 SOQ40932.1 AGBW02002272 OWR55647.1 DS235882 EEB20093.1 GBXI01007108 JAD07184.1 GBXI01012836 GBXI01008793 JAD01456.1 JAD05499.1 KY849642 GAKP01001123 ATY74523.1 JAC57829.1 GL888378 EGI62032.1 KQ976738 KYM75695.1 KQ981414 KYN41879.1 ADTU01005818 ADTU01005819 ADTU01005820 GDHF01019803 GDHF01009501 JAI32511.1 JAI42813.1 GBYB01001715 JAG71482.1 KK107235 QOIP01000009 EZA54901.1 RLU19115.1 GBYB01001713 JAG71480.1 NNAY01001302 OXU24428.1 KQ982691 KYQ52091.1 KA645370 AFP59999.1 JRES01000418 KNC31437.1 KQ971338 EFA02729.1 JXUM01044305 JXUM01044306 JXUM01044307 JXUM01044308 JXUM01044309 JXUM01044310 JXUM01044311 JXUM01044312 KQ561395 KXJ78719.1 JXUM01043670 JXUM01043671 KQ561371 KXJ78816.1 GL767121 EFZ14056.1 CH477522 EAT39584.1 CH902620 EDV32237.1 ATLV01015024 KE524999 KFB39951.1 AXCM01002128 KK853336 KDR08337.1 ADMH02002133 ETN58461.1 GGFK01002636 MBW35957.1 AAAB01008839 EAA45250.4 APCN01004583 GFDL01002429 JAV32616.1 CH954177 EDV58434.1 GGFJ01002997 MBW52138.1 GALX01004992 JAB63474.1 CH963857 EDW76669.1 GGFK01002639 MBW35960.1 KQ434783 KZC04865.1 CH379060 EAL33391.1 KRT03789.1 AY052113 AAK93537.1 GGFM01001231 MBW21982.1 GGFM01001267 MBW22018.1 CM002910 KMY89371.1 GEZM01087692 JAV58531.1 CM000361 EDX04427.1 CM000157 EDW88714.1 AE014134 BT150417 AAO41181.1 AHB32099.1 OUUW01000010 SPP85635.1 KB632374 ERL93810.1 CH916368 EDW03116.1 GFDF01000508 JAV13576.1 CH940654 EDW57835.1 GANO01002306 JAB57565.1 CP012523 ALC39044.1 ALC40218.1 KQ977141 KYN05399.1 UFQT01000426 SSX24216.1 GARF01000053 JAC88899.1 CH933807 EDW12241.1 KRG03163.1 KRG03164.1 CCAG010007529 KQ414705 KOC63205.1 NEVH01005883 PNF38654.1 GEBQ01024653 JAT15324.1 JXJN01023980 JXJN01023981 AJWK01017953 AJWK01017954 GECZ01028282 JAS41487.1 GL436038 EFN72387.1 KF828797 AIN44114.1 AK418035 BAN21250.1 GBHO01038197 GBHO01010744 GBHO01010743 GBHO01001716 GBHO01001707 GDHC01006544 JAG05407.1 JAG32860.1 JAG32861.1 JAG41888.1 JAG41897.1 JAQ12085.1 GBHO01038204 GBHO01038203 GBHO01038201 GBHO01038200 GBHO01038198 GDHC01017926 GDHC01006925 JAG05400.1 JAG05401.1 JAG05403.1 JAG05404.1 JAG05406.1 JAQ00703.1 JAQ11704.1 CVRI01000020 CRK90572.1 CH480818 EDW52330.1 GALX01004991 JAB63475.1
ADZ56944.1 GAIX01006768 JAA85792.1 KQ459592 KPI96829.1 KQ460636 KPJ13379.1 NWSH01000443 PCG76396.1 PCG76395.1 ODYU01002885 SOQ40932.1 AGBW02002272 OWR55647.1 DS235882 EEB20093.1 GBXI01007108 JAD07184.1 GBXI01012836 GBXI01008793 JAD01456.1 JAD05499.1 KY849642 GAKP01001123 ATY74523.1 JAC57829.1 GL888378 EGI62032.1 KQ976738 KYM75695.1 KQ981414 KYN41879.1 ADTU01005818 ADTU01005819 ADTU01005820 GDHF01019803 GDHF01009501 JAI32511.1 JAI42813.1 GBYB01001715 JAG71482.1 KK107235 QOIP01000009 EZA54901.1 RLU19115.1 GBYB01001713 JAG71480.1 NNAY01001302 OXU24428.1 KQ982691 KYQ52091.1 KA645370 AFP59999.1 JRES01000418 KNC31437.1 KQ971338 EFA02729.1 JXUM01044305 JXUM01044306 JXUM01044307 JXUM01044308 JXUM01044309 JXUM01044310 JXUM01044311 JXUM01044312 KQ561395 KXJ78719.1 JXUM01043670 JXUM01043671 KQ561371 KXJ78816.1 GL767121 EFZ14056.1 CH477522 EAT39584.1 CH902620 EDV32237.1 ATLV01015024 KE524999 KFB39951.1 AXCM01002128 KK853336 KDR08337.1 ADMH02002133 ETN58461.1 GGFK01002636 MBW35957.1 AAAB01008839 EAA45250.4 APCN01004583 GFDL01002429 JAV32616.1 CH954177 EDV58434.1 GGFJ01002997 MBW52138.1 GALX01004992 JAB63474.1 CH963857 EDW76669.1 GGFK01002639 MBW35960.1 KQ434783 KZC04865.1 CH379060 EAL33391.1 KRT03789.1 AY052113 AAK93537.1 GGFM01001231 MBW21982.1 GGFM01001267 MBW22018.1 CM002910 KMY89371.1 GEZM01087692 JAV58531.1 CM000361 EDX04427.1 CM000157 EDW88714.1 AE014134 BT150417 AAO41181.1 AHB32099.1 OUUW01000010 SPP85635.1 KB632374 ERL93810.1 CH916368 EDW03116.1 GFDF01000508 JAV13576.1 CH940654 EDW57835.1 GANO01002306 JAB57565.1 CP012523 ALC39044.1 ALC40218.1 KQ977141 KYN05399.1 UFQT01000426 SSX24216.1 GARF01000053 JAC88899.1 CH933807 EDW12241.1 KRG03163.1 KRG03164.1 CCAG010007529 KQ414705 KOC63205.1 NEVH01005883 PNF38654.1 GEBQ01024653 JAT15324.1 JXJN01023980 JXJN01023981 AJWK01017953 AJWK01017954 GECZ01028282 JAS41487.1 GL436038 EFN72387.1 KF828797 AIN44114.1 AK418035 BAN21250.1 GBHO01038197 GBHO01010744 GBHO01010743 GBHO01001716 GBHO01001707 GDHC01006544 JAG05407.1 JAG32860.1 JAG32861.1 JAG41888.1 JAG41897.1 JAQ12085.1 GBHO01038204 GBHO01038203 GBHO01038201 GBHO01038200 GBHO01038198 GDHC01017926 GDHC01006925 JAG05400.1 JAG05401.1 JAG05403.1 JAG05404.1 JAG05406.1 JAQ00703.1 JAQ11704.1 CVRI01000020 CRK90572.1 CH480818 EDW52330.1 GALX01004991 JAB63475.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000009046
+ More
UP000002358 UP000007755 UP000192223 UP000078540 UP000078541 UP000005205 UP000053097 UP000279307 UP000215335 UP000075809 UP000095301 UP000037069 UP000007266 UP000069940 UP000249989 UP000008820 UP000007801 UP000075900 UP000075881 UP000075885 UP000030765 UP000095300 UP000075883 UP000027135 UP000000673 UP000076408 UP000069272 UP000076407 UP000075902 UP000075903 UP000075882 UP000007062 UP000075840 UP000008711 UP000007798 UP000076502 UP000075880 UP000001819 UP000075920 UP000192221 UP000000304 UP000002282 UP000000803 UP000268350 UP000030742 UP000001070 UP000008792 UP000092553 UP000078542 UP000091820 UP000092443 UP000009192 UP000078200 UP000092445 UP000092444 UP000053825 UP000235965 UP000092460 UP000092461 UP000000311 UP000183832 UP000001292
UP000002358 UP000007755 UP000192223 UP000078540 UP000078541 UP000005205 UP000053097 UP000279307 UP000215335 UP000075809 UP000095301 UP000037069 UP000007266 UP000069940 UP000249989 UP000008820 UP000007801 UP000075900 UP000075881 UP000075885 UP000030765 UP000095300 UP000075883 UP000027135 UP000000673 UP000076408 UP000069272 UP000076407 UP000075902 UP000075903 UP000075882 UP000007062 UP000075840 UP000008711 UP000007798 UP000076502 UP000075880 UP000001819 UP000075920 UP000192221 UP000000304 UP000002282 UP000000803 UP000268350 UP000030742 UP000001070 UP000008792 UP000092553 UP000078542 UP000091820 UP000092443 UP000009192 UP000078200 UP000092445 UP000092444 UP000053825 UP000235965 UP000092460 UP000092461 UP000000311 UP000183832 UP000001292
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9J6T4
F2YDQ6
S4PWH7
A0A194PVN9
A0A0N0PCC8
A0A2A4JX93
+ More
A0A2A4JWK0 A0A2H1VKG2 A0A212FPJ2 E0W387 A0A0A1X7A6 A0A0A1X2C7 K7ILU8 A0A034WT57 F4WUW8 A0A1W4XHH8 A0A195AUJ8 A0A151JZL1 A0A158P0V0 A0A0K8V0X6 A0A0C9R177 A0A026WGT2 A0A0C9PL33 A0A232F1V9 A0A151WWP2 T1PB46 A0A0L0CGW6 D2A232 A0A182G655 A0A182G5I4 E9IZD3 Q16Y84 B3MPB8 A0A1S4FK15 A0A182RGB5 A0A182K839 A0A182P7F0 A0A084VPQ7 A0A1I8P8P9 A0A182MVP0 A0A067QV98 W5J3P6 A0A182YG04 A0A182F592 A0A2M4A571 A0A182XFM9 A0A182TXZ8 A0A182UP98 A0A182L5H1 Q7PFM9 A0A182IE85 A0A1Q3FYJ3 B3N8L1 A0A2M4BH59 V5GHI9 B4MX30 A0A2M4A5F4 A0A154NYW5 A0A182JJD9 Q29NF1 Q960D7 A0A2M3Z0C2 A0A182VUV0 A0A2M3Z0Q0 A0A1W4UUD9 A0A0J9QZL2 A0A1Y1KDU5 B4Q8A5 B4NYQ0 Q9VL61 A0A3B0JU51 U4UUH1 B4JCG2 A0A1L8E4I7 B4M9F9 U5EUI7 A0A0M5J908 A0A195CZ41 A0A336M1M3 A0A1A9WHI6 A0A1A9XMQ4 A0A0U9HYG1 B4KH30 A0A1A9VE61 A0A1B0ABB5 A0A1B0G0H7 A0A0L7QX80 A0A2J7RCU0 A0A1B6KVQ4 A0A1B0C1D6 A0A1B0GJ02 A0A1B6EU87 E2A2I4 A0A161ANB1 R4WKD2 A0A0A9YMK9 A0A0A9WFV8 A0A1J1HT67 B4HWD5 V5G0H5
A0A2A4JWK0 A0A2H1VKG2 A0A212FPJ2 E0W387 A0A0A1X7A6 A0A0A1X2C7 K7ILU8 A0A034WT57 F4WUW8 A0A1W4XHH8 A0A195AUJ8 A0A151JZL1 A0A158P0V0 A0A0K8V0X6 A0A0C9R177 A0A026WGT2 A0A0C9PL33 A0A232F1V9 A0A151WWP2 T1PB46 A0A0L0CGW6 D2A232 A0A182G655 A0A182G5I4 E9IZD3 Q16Y84 B3MPB8 A0A1S4FK15 A0A182RGB5 A0A182K839 A0A182P7F0 A0A084VPQ7 A0A1I8P8P9 A0A182MVP0 A0A067QV98 W5J3P6 A0A182YG04 A0A182F592 A0A2M4A571 A0A182XFM9 A0A182TXZ8 A0A182UP98 A0A182L5H1 Q7PFM9 A0A182IE85 A0A1Q3FYJ3 B3N8L1 A0A2M4BH59 V5GHI9 B4MX30 A0A2M4A5F4 A0A154NYW5 A0A182JJD9 Q29NF1 Q960D7 A0A2M3Z0C2 A0A182VUV0 A0A2M3Z0Q0 A0A1W4UUD9 A0A0J9QZL2 A0A1Y1KDU5 B4Q8A5 B4NYQ0 Q9VL61 A0A3B0JU51 U4UUH1 B4JCG2 A0A1L8E4I7 B4M9F9 U5EUI7 A0A0M5J908 A0A195CZ41 A0A336M1M3 A0A1A9WHI6 A0A1A9XMQ4 A0A0U9HYG1 B4KH30 A0A1A9VE61 A0A1B0ABB5 A0A1B0G0H7 A0A0L7QX80 A0A2J7RCU0 A0A1B6KVQ4 A0A1B0C1D6 A0A1B0GJ02 A0A1B6EU87 E2A2I4 A0A161ANB1 R4WKD2 A0A0A9YMK9 A0A0A9WFV8 A0A1J1HT67 B4HWD5 V5G0H5
PDB
6FFC
E-value=2.23755e-44,
Score=451
Ontologies
KEGG
GO
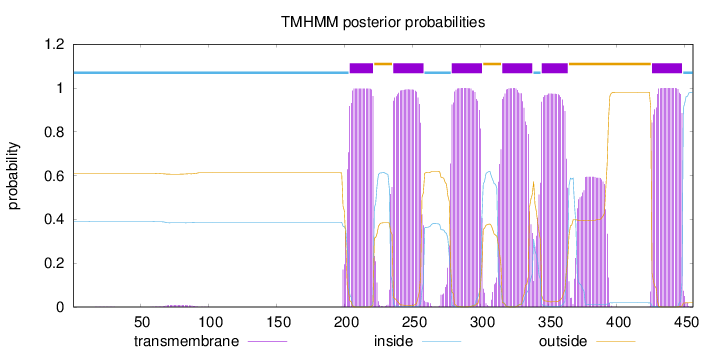
Topology
Length:
456
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
145.79527
Exp number, first 60 AAs:
0.00428
Total prob of N-in:
0.39084
inside
1 - 203
TMhelix
204 - 221
outside
222 - 235
TMhelix
236 - 258
inside
259 - 278
TMhelix
279 - 301
outside
302 - 315
TMhelix
316 - 338
inside
339 - 344
TMhelix
345 - 364
outside
365 - 425
TMhelix
426 - 448
inside
449 - 456
Population Genetic Test Statistics
Pi
22.942309
Theta
23.255576
Tajima's D
0.118304
CLR
0.656897
CSRT
0.408529573521324
Interpretation
Uncertain
