Gene
KWMTBOMO07442
Pre Gene Modal
BGIBMGA005230
Annotation
PREDICTED:_dnaJ_homolog_subfamily_C_member_10_isoform_X1_[Bombyx_mori]
Full name
DnaJ homolog subfamily C member 10
Location in the cell
PlasmaMembrane Reliability : 2.147
Sequence
CDS
ATGCAACCAATAATGTTAAAACTAGTATTTATCGTGATAGTCCTTATCGAAGCGGTTAACCTCACAGACGTATCATATTATGAAATTCTAGGGATAACTAAACAGGCATCGACTCAGGAAATAAAACAGGCGTATAAAAAATTAGTCGTAAAGTTTCACCCTGATAAAAATCCTAATGAAGCCAAACAAGAAAAATTTCTTAAAATAACTGAAGCCTATGAAACCTTGAAGGATCCAGAGAAAAGGCGCAATTATGATTTGTATGGATCATACACAACTTATTCAAGAAAATATGATTATAAATCTCAGTCGGAATATGATAATTTGTACTATAAAGGACTGTACCATAATGATCCATTTGTTGACACTTTATCTGGATCAAGTTTCTACAATTACCTGAATGAAGGATTTCACTTTATAAATTTTTACTCACCATTTTGTCCCCCCTGTCAAAATATTGCAGATCATTGGAAGAAGTTAGCAGAAATATATAAGGGCATTGTGAAAGTCGGTGCTGTTAACTGCAAATACCATAACTCATTTTGTTATAACTCAATGCGAATTGGCAGCTATCCAACTCTATTATTTTACCCTAATGGTAAAAGTGGAAACTATTTATATTACCGAGGCGACAGGACTTTCGAGGCTCTAGATGATTTTGTAATGACATACCTAAACAGTCATGTGCATGTGCCTACCGTCTCTCAACTGCGAAATACCGACAGGCCAATCGCCTACGTTCTCGGTGCTAATAGAATAGACAGGAGTGCCTTAACAAGGATTGCATATCGATTGAACGGCCTTGTAGCAATCGCTATCGTAGAAGACGAGAGCTTGAGAGATAAATTATCCGAAAGTGACTACACAACGATCGTGTTCAAATACAAAGGTTTTATTAAAGACATAGAGAGTACAAATGAGGAAGATGTTCTAAAAGATATAGTGGCTGCCTTGCCGAAGATCGAGCTAATTGATCCAGAAAAGTTGAAGAACATACGTAATAAGTTACGTAGAGGAGAACAAACGCCTTGGGTCCTGTATTTCTCCACCAAGGGAAGTGACAGACTCGTTCTACATCAAATGAGAATGTCGTTCCCTAATTTAAATTTTGGCGAGATCGATTGCGACGTATGGGCGGAGCTGTGCTCGTCGCTGCACGTGGAGGCGGCGCCGGCGTGGGCGGTCCTGAAGCGCGGCGGCGCCTACCAGCGGGCGTACGCGCCGGCGGACGCACGCACCTTCGTCCGCACCGCCGCCGCCGCCCTCAGCCTGCACACGCTCTCCGCTTCCGACCTCAACAAGATCTTGGAAGGGGATGTCGGCGTGTGGGTGCTGCTGGTAGTTCCGCACGGATCGCCGTGGGACAGCATGGCCGGACCCTTCACTCAGACCAGCATGCATTTCAACAACGACGAAAACGTCAGTTTCGGCATAATGATATGCACGGCAAGCACCGACCAGCACTGCCGACGAGTGGCGCCGGAGAAGCCCGTCGTGCTTCTGCAGAACTTCACCAGGCGGCACACGTACCGCGGCGAGCTTAACGAGAGGGAGCTTTTGGAGTACATCAATATGCTGAGAGACAGTGAAGATCTTGAGTTGACGGAGAATCAAGTTTTAGAGATATCGGATCCGAGCCGCGAGCACGCGTGGCTGGTGGCGTACCTGCCCGCGCACTGCGGCGCCTTCTGCGACGACCTCGCCCACCACTGGATGATCGTTGCCAGCAAGCTGCGACCTCTGACTTTTGTTCGAGTTGGAATGTTGCAATGTGCAAAAATAAAGAGCGGTTTCTGTACAAACATTCGATCAGCCACGGCAAGACTCTATCCTATAGCGTCTGGACAACATTATAGCGTCAGTCTACAGCATTTATCGCAAGCTCCGTACATACTGGAATGGGCGTTACAGTTCATAGACGATTCGGTTGTGAAACTCAACTGGAAACTGTTCGCTAAGACCGTTATTGCCGAGGAACTGAACCCGACGAGCGGCAACAAACCTTGGCTGGTCTATTTCCACTCTCCCAGATGTTACCGCTGCTACGAGATGTACGCTGACTTTGCTATAGCCGGTATTCTACTGAACAACGCGGTGCAGCTCGGCAAAGTCAATTGCCTGAACGAGCGCGGCCTCTGCCAACACGAGCACATCACAAGCTATCCGTCGCTCAAGCTGTACCTCGGCAGGAACAGCCGCCAGCGCTTCAGCAGCGTCGTGACACTGCAGGTCAGAGACCACAAGAGCATTCTAGAAGAAATCGAACCTCATTTAAGGAGATACGACGCCAGTCTGCTGGCCACCCGCGACGGTGCTGCCAGTAATAGCATCAGACACGACGAATTTTAA
Protein
MQPIMLKLVFIVIVLIEAVNLTDVSYYEILGITKQASTQEIKQAYKKLVVKFHPDKNPNEAKQEKFLKITEAYETLKDPEKRRNYDLYGSYTTYSRKYDYKSQSEYDNLYYKGLYHNDPFVDTLSGSSFYNYLNEGFHFINFYSPFCPPCQNIADHWKKLAEIYKGIVKVGAVNCKYHNSFCYNSMRIGSYPTLLFYPNGKSGNYLYYRGDRTFEALDDFVMTYLNSHVHVPTVSQLRNTDRPIAYVLGANRIDRSALTRIAYRLNGLVAIAIVEDESLRDKLSESDYTTIVFKYKGFIKDIESTNEEDVLKDIVAALPKIELIDPEKLKNIRNKLRRGEQTPWVLYFSTKGSDRLVLHQMRMSFPNLNFGEIDCDVWAELCSSLHVEAAPAWAVLKRGGAYQRAYAPADARTFVRTAAAALSLHTLSASDLNKILEGDVGVWVLLVVPHGSPWDSMAGPFTQTSMHFNNDENVSFGIMICTASTDQHCRRVAPEKPVVLLQNFTRRHTYRGELNERELLEYINMLRDSEDLELTENQVLEISDPSREHAWLVAYLPAHCGAFCDDLAHHWMIVASKLRPLTFVRVGMLQCAKIKSGFCTNIRSATARLYPIASGQHYSVSLQHLSQAPYILEWALQFIDDSVVKLNWKLFAKTVIAEELNPTSGNKPWLVYFHSPRCYRCYEMYADFAIAGILLNNAVQLGKVNCLNERGLCQHEHITSYPSLKLYLGRNSRQRFSSVVTLQVRDHKSILEEIEPHLRRYDASLLATRDGAASNSIRHDEF
Summary
Description
Endoplasmic reticulum disulfide reductase involved both in the correct folding of proteins and degradation of misfolded proteins. Required for efficient folding of proteins in the endoplasmic reticulum by catalyzing the removal of non-native disulfide bonds formed during the folding of proteins. Also involved in endoplasmic reticulum-associated degradation (ERAD) by reducing incorrect disulfide bonds in misfolded glycoproteins (By similarity).
Endoplasmic reticulum disulfide reductase involved both in the correct folding of proteins and degradation of misfolded proteins.
Endoplasmic reticulum disulfide reductase involved both in the correct folding of proteins and degradation of misfolded proteins.
Keywords
Disulfide bond
Endoplasmic reticulum
Glycoprotein
Oxidoreductase
Redox-active center
Repeat
Signal
Feature
chain DnaJ homolog subfamily C member 10
Uniprot
A0A2A4JY20
A0A3S2P550
A0A194PU39
A0A2H1VKE8
A0A0N1I782
A0A212FFB3
+ More
A0A1W4XRD8 N6U1H5 D6WB87 K7IUJ8 A0A067R8R7 A0A0T6AXX5 A0A232EVB6 A0A1B6K1C2 E9HSV1 A0A0C9RUD2 L7M9R9 A0A224YLI3 A0A131YX50 A0A1S3ICH4 A0A131XCL6 V4CLN4 A0A1B6D3Y5 A0A0A9WTW6 A0A0P6JMH9 A0A195CP40 K1R407 A0A164XM53 E2C8F2 A0A0P5E587 T1ICK1 A7RXE0 A0A210QKP4 A0A0P5W9U5 U4UEY1 A0A0P4VZ11 A0A091J2M0 A0A158NIB7 A0A091G042 Q6NRT6 A0A1L8EPM5 A0A195EFX2 R0LS95 A0A091PT94 U3J188 A0A218UPK8 A0A091HR19 F4WGA6 A0A093D0G1 A0A091HEW0 A0A224X7Y8 A0A2T7Q1M0 G1N9Z6 A0A091NRI4 A0A093NIJ9 A0A093QSW7 A0A093EV94 A0A091PMM1 U3JLW4 A0A093IVM4 A0A0A0AL06 T1IPH3 A0A0P5XH85 A0A091QP43 A0A087QGY3 T1ED98 A0A2I0UPW3 A0A091VNZ0 H0ZH31 A0A0P4YVH1 E1BRA6 A0A099ZAI7 A0A0P5UBM5 A0A1L8EWF7 A0A091KIA8 A0A091M795 A0A093GH98 A0A091EPS8 A0A1V4L0K1 A0A3B1J415 A0A091UV72 A0A2I0LXI9 A0A3L8SEP1 F7FJY1 A0A0B8RZ19 A0A093HHB7 B7PNU9 A0A151LZP1 A0A3B4BQ05 G3VP55 K7F258 W5UIN2 R7TG96 A0A2D0R3S8 A0A2D0R5C2 F7E9S8
A0A1W4XRD8 N6U1H5 D6WB87 K7IUJ8 A0A067R8R7 A0A0T6AXX5 A0A232EVB6 A0A1B6K1C2 E9HSV1 A0A0C9RUD2 L7M9R9 A0A224YLI3 A0A131YX50 A0A1S3ICH4 A0A131XCL6 V4CLN4 A0A1B6D3Y5 A0A0A9WTW6 A0A0P6JMH9 A0A195CP40 K1R407 A0A164XM53 E2C8F2 A0A0P5E587 T1ICK1 A7RXE0 A0A210QKP4 A0A0P5W9U5 U4UEY1 A0A0P4VZ11 A0A091J2M0 A0A158NIB7 A0A091G042 Q6NRT6 A0A1L8EPM5 A0A195EFX2 R0LS95 A0A091PT94 U3J188 A0A218UPK8 A0A091HR19 F4WGA6 A0A093D0G1 A0A091HEW0 A0A224X7Y8 A0A2T7Q1M0 G1N9Z6 A0A091NRI4 A0A093NIJ9 A0A093QSW7 A0A093EV94 A0A091PMM1 U3JLW4 A0A093IVM4 A0A0A0AL06 T1IPH3 A0A0P5XH85 A0A091QP43 A0A087QGY3 T1ED98 A0A2I0UPW3 A0A091VNZ0 H0ZH31 A0A0P4YVH1 E1BRA6 A0A099ZAI7 A0A0P5UBM5 A0A1L8EWF7 A0A091KIA8 A0A091M795 A0A093GH98 A0A091EPS8 A0A1V4L0K1 A0A3B1J415 A0A091UV72 A0A2I0LXI9 A0A3L8SEP1 F7FJY1 A0A0B8RZ19 A0A093HHB7 B7PNU9 A0A151LZP1 A0A3B4BQ05 G3VP55 K7F258 W5UIN2 R7TG96 A0A2D0R3S8 A0A2D0R5C2 F7E9S8
EC Number
1.8.4.-
Pubmed
26354079
22118469
23537049
18362917
19820115
20075255
+ More
24845553 28648823 21292972 25576852 28797301 26830274 28049606 23254933 25401762 22992520 20798317 17615350 28812685 21347285 27762356 23749191 21719571 20838655 20360741 15592404 25329095 23371554 30282656 18464734 25476704 22293439 21709235 17381049 23127152 17495919
24845553 28648823 21292972 25576852 28797301 26830274 28049606 23254933 25401762 22992520 20798317 17615350 28812685 21347285 27762356 23749191 21719571 20838655 20360741 15592404 25329095 23371554 30282656 18464734 25476704 22293439 21709235 17381049 23127152 17495919
EMBL
NWSH01000443
PCG76393.1
RSAL01000020
RVE52633.1
KQ459592
KPI96832.1
+ More
ODYU01003032 SOQ41295.1 KQ460636 KPJ13373.1 AGBW02008839 OWR52419.1 APGK01043268 APGK01043269 APGK01043270 KB741014 ENN75400.1 KQ971312 EEZ97922.2 AAZX01002625 KK852809 KDR15988.1 LJIG01022541 KRT80031.1 NNAY01002021 OXU22293.1 GECU01002447 JAT05260.1 GL732758 EFX65186.1 GBYB01012425 JAG82192.1 GACK01005175 JAA59859.1 GFPF01007351 MAA18497.1 GEDV01004794 JAP83763.1 GEFH01004716 JAP63865.1 KB200084 ESP03215.1 GEDC01016920 GEDC01004515 JAS20378.1 JAS32783.1 GBHO01032738 JAG10866.1 GDIQ01002824 JAN91913.1 KQ977481 KYN02410.1 JH817910 EKC28571.1 LRGB01000966 KZS14374.1 GL453666 EFN75787.1 GDIP01151373 JAJ72029.1 ACPB03013479 DS469550 EDO43758.1 NEDP02003178 OWF49324.1 GDIP01102381 JAM01334.1 KB632328 ERL92534.1 GDRN01101528 JAI58356.1 KK501437 KFP14852.1 ADTU01016302 ADTU01016303 KL447622 KFO75293.1 BC070632 CM004483 OCT61219.1 KQ978957 KYN27061.1 KB742873 EOB03273.1 KK678051 KFQ10551.1 ADON01053152 ADON01053153 MUZQ01000205 OWK55350.1 KL217737 KFO98336.1 GL888128 EGI66873.1 KL474897 KFV20258.1 KL535944 KFO94411.1 GFTR01007960 JAW08466.1 PZQS01000001 PVD39550.1 KL393317 KFP92393.1 KL224607 KFW61830.1 KL431002 KFW91621.1 KK382500 KFV49217.1 KK661632 KFQ08586.1 AGTO01000451 KK563429 KFW02829.1 KL872189 KGL94721.1 JH431262 GDIP01073902 JAM29813.1 KK702751 KFQ28376.1 KL225576 KFM00487.1 AMQM01006708 KB097502 ESN95836.1 KZ505661 PKU48075.1 KK734218 KFR05152.1 ABQF01010254 GDIP01223064 JAJ00338.1 AADN05000350 KL890656 KGL77983.1 GDIP01115385 JAL88329.1 CM004482 OCT63684.1 KK746325 KFP40339.1 KK524674 KFP67673.1 KL215731 KFV66297.1 KK718916 KFO59913.1 LSYS01000429 OPJ90162.1 KL410237 KFQ94854.1 AKCR02000066 PKK22148.1 QUSF01000024 RLW00998.1 GBSH01000882 JAG68143.1 KL206406 KFV82033.1 ABJB010074310 DS754779 EEC08271.1 AKHW03006853 KYO17745.1 AEFK01112961 AEFK01112962 AGCU01076169 AGCU01076170 AGCU01076171 JT415868 AHH41846.1 AMQN01002771 KB310004 ELT92793.1
ODYU01003032 SOQ41295.1 KQ460636 KPJ13373.1 AGBW02008839 OWR52419.1 APGK01043268 APGK01043269 APGK01043270 KB741014 ENN75400.1 KQ971312 EEZ97922.2 AAZX01002625 KK852809 KDR15988.1 LJIG01022541 KRT80031.1 NNAY01002021 OXU22293.1 GECU01002447 JAT05260.1 GL732758 EFX65186.1 GBYB01012425 JAG82192.1 GACK01005175 JAA59859.1 GFPF01007351 MAA18497.1 GEDV01004794 JAP83763.1 GEFH01004716 JAP63865.1 KB200084 ESP03215.1 GEDC01016920 GEDC01004515 JAS20378.1 JAS32783.1 GBHO01032738 JAG10866.1 GDIQ01002824 JAN91913.1 KQ977481 KYN02410.1 JH817910 EKC28571.1 LRGB01000966 KZS14374.1 GL453666 EFN75787.1 GDIP01151373 JAJ72029.1 ACPB03013479 DS469550 EDO43758.1 NEDP02003178 OWF49324.1 GDIP01102381 JAM01334.1 KB632328 ERL92534.1 GDRN01101528 JAI58356.1 KK501437 KFP14852.1 ADTU01016302 ADTU01016303 KL447622 KFO75293.1 BC070632 CM004483 OCT61219.1 KQ978957 KYN27061.1 KB742873 EOB03273.1 KK678051 KFQ10551.1 ADON01053152 ADON01053153 MUZQ01000205 OWK55350.1 KL217737 KFO98336.1 GL888128 EGI66873.1 KL474897 KFV20258.1 KL535944 KFO94411.1 GFTR01007960 JAW08466.1 PZQS01000001 PVD39550.1 KL393317 KFP92393.1 KL224607 KFW61830.1 KL431002 KFW91621.1 KK382500 KFV49217.1 KK661632 KFQ08586.1 AGTO01000451 KK563429 KFW02829.1 KL872189 KGL94721.1 JH431262 GDIP01073902 JAM29813.1 KK702751 KFQ28376.1 KL225576 KFM00487.1 AMQM01006708 KB097502 ESN95836.1 KZ505661 PKU48075.1 KK734218 KFR05152.1 ABQF01010254 GDIP01223064 JAJ00338.1 AADN05000350 KL890656 KGL77983.1 GDIP01115385 JAL88329.1 CM004482 OCT63684.1 KK746325 KFP40339.1 KK524674 KFP67673.1 KL215731 KFV66297.1 KK718916 KFO59913.1 LSYS01000429 OPJ90162.1 KL410237 KFQ94854.1 AKCR02000066 PKK22148.1 QUSF01000024 RLW00998.1 GBSH01000882 JAG68143.1 KL206406 KFV82033.1 ABJB010074310 DS754779 EEC08271.1 AKHW03006853 KYO17745.1 AEFK01112961 AEFK01112962 AGCU01076169 AGCU01076170 AGCU01076171 JT415868 AHH41846.1 AMQN01002771 KB310004 ELT92793.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
UP000192223
+ More
UP000019118 UP000007266 UP000002358 UP000027135 UP000215335 UP000000305 UP000085678 UP000030746 UP000078542 UP000005408 UP000076858 UP000008237 UP000015103 UP000001593 UP000242188 UP000030742 UP000053119 UP000005205 UP000053760 UP000186698 UP000078492 UP000016666 UP000197619 UP000054308 UP000007755 UP000245119 UP000001645 UP000054081 UP000016665 UP000053858 UP000053286 UP000015101 UP000053605 UP000007754 UP000000539 UP000053641 UP000053875 UP000052976 UP000190648 UP000018467 UP000053283 UP000053872 UP000276834 UP000002279 UP000053584 UP000001555 UP000050525 UP000261440 UP000007648 UP000007267 UP000014760 UP000221080 UP000002280
UP000019118 UP000007266 UP000002358 UP000027135 UP000215335 UP000000305 UP000085678 UP000030746 UP000078542 UP000005408 UP000076858 UP000008237 UP000015103 UP000001593 UP000242188 UP000030742 UP000053119 UP000005205 UP000053760 UP000186698 UP000078492 UP000016666 UP000197619 UP000054308 UP000007755 UP000245119 UP000001645 UP000054081 UP000016665 UP000053858 UP000053286 UP000015101 UP000053605 UP000007754 UP000000539 UP000053641 UP000053875 UP000052976 UP000190648 UP000018467 UP000053283 UP000053872 UP000276834 UP000002279 UP000053584 UP000001555 UP000050525 UP000261440 UP000007648 UP000007267 UP000014760 UP000221080 UP000002280
Pfam
Interpro
IPR001623
DnaJ_domain
+ More
IPR017937 Thioredoxin_CS
IPR013766 Thioredoxin_domain
IPR036869 J_dom_sf
IPR036249 Thioredoxin-like_sf
IPR018253 DnaJ_domain_CS
IPR011701 MFS
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR005829 Sugar_transporter_CS
IPR001431 Pept_M16_Zn_BS
IPR011765 Pept_M16_N
IPR007863 Peptidase_M16_C
IPR032632 Peptidase_M16_M
IPR011249 Metalloenz_LuxS/M16
IPR035674 ERdj5_TRX_C
IPR021170 ERdj5
IPR035673 ERdj5_TRX_N
IPR017937 Thioredoxin_CS
IPR013766 Thioredoxin_domain
IPR036869 J_dom_sf
IPR036249 Thioredoxin-like_sf
IPR018253 DnaJ_domain_CS
IPR011701 MFS
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR005829 Sugar_transporter_CS
IPR001431 Pept_M16_Zn_BS
IPR011765 Pept_M16_N
IPR007863 Peptidase_M16_C
IPR032632 Peptidase_M16_M
IPR011249 Metalloenz_LuxS/M16
IPR035674 ERdj5_TRX_C
IPR021170 ERdj5
IPR035673 ERdj5_TRX_N
Gene 3D
ProteinModelPortal
A0A2A4JY20
A0A3S2P550
A0A194PU39
A0A2H1VKE8
A0A0N1I782
A0A212FFB3
+ More
A0A1W4XRD8 N6U1H5 D6WB87 K7IUJ8 A0A067R8R7 A0A0T6AXX5 A0A232EVB6 A0A1B6K1C2 E9HSV1 A0A0C9RUD2 L7M9R9 A0A224YLI3 A0A131YX50 A0A1S3ICH4 A0A131XCL6 V4CLN4 A0A1B6D3Y5 A0A0A9WTW6 A0A0P6JMH9 A0A195CP40 K1R407 A0A164XM53 E2C8F2 A0A0P5E587 T1ICK1 A7RXE0 A0A210QKP4 A0A0P5W9U5 U4UEY1 A0A0P4VZ11 A0A091J2M0 A0A158NIB7 A0A091G042 Q6NRT6 A0A1L8EPM5 A0A195EFX2 R0LS95 A0A091PT94 U3J188 A0A218UPK8 A0A091HR19 F4WGA6 A0A093D0G1 A0A091HEW0 A0A224X7Y8 A0A2T7Q1M0 G1N9Z6 A0A091NRI4 A0A093NIJ9 A0A093QSW7 A0A093EV94 A0A091PMM1 U3JLW4 A0A093IVM4 A0A0A0AL06 T1IPH3 A0A0P5XH85 A0A091QP43 A0A087QGY3 T1ED98 A0A2I0UPW3 A0A091VNZ0 H0ZH31 A0A0P4YVH1 E1BRA6 A0A099ZAI7 A0A0P5UBM5 A0A1L8EWF7 A0A091KIA8 A0A091M795 A0A093GH98 A0A091EPS8 A0A1V4L0K1 A0A3B1J415 A0A091UV72 A0A2I0LXI9 A0A3L8SEP1 F7FJY1 A0A0B8RZ19 A0A093HHB7 B7PNU9 A0A151LZP1 A0A3B4BQ05 G3VP55 K7F258 W5UIN2 R7TG96 A0A2D0R3S8 A0A2D0R5C2 F7E9S8
A0A1W4XRD8 N6U1H5 D6WB87 K7IUJ8 A0A067R8R7 A0A0T6AXX5 A0A232EVB6 A0A1B6K1C2 E9HSV1 A0A0C9RUD2 L7M9R9 A0A224YLI3 A0A131YX50 A0A1S3ICH4 A0A131XCL6 V4CLN4 A0A1B6D3Y5 A0A0A9WTW6 A0A0P6JMH9 A0A195CP40 K1R407 A0A164XM53 E2C8F2 A0A0P5E587 T1ICK1 A7RXE0 A0A210QKP4 A0A0P5W9U5 U4UEY1 A0A0P4VZ11 A0A091J2M0 A0A158NIB7 A0A091G042 Q6NRT6 A0A1L8EPM5 A0A195EFX2 R0LS95 A0A091PT94 U3J188 A0A218UPK8 A0A091HR19 F4WGA6 A0A093D0G1 A0A091HEW0 A0A224X7Y8 A0A2T7Q1M0 G1N9Z6 A0A091NRI4 A0A093NIJ9 A0A093QSW7 A0A093EV94 A0A091PMM1 U3JLW4 A0A093IVM4 A0A0A0AL06 T1IPH3 A0A0P5XH85 A0A091QP43 A0A087QGY3 T1ED98 A0A2I0UPW3 A0A091VNZ0 H0ZH31 A0A0P4YVH1 E1BRA6 A0A099ZAI7 A0A0P5UBM5 A0A1L8EWF7 A0A091KIA8 A0A091M795 A0A093GH98 A0A091EPS8 A0A1V4L0K1 A0A3B1J415 A0A091UV72 A0A2I0LXI9 A0A3L8SEP1 F7FJY1 A0A0B8RZ19 A0A093HHB7 B7PNU9 A0A151LZP1 A0A3B4BQ05 G3VP55 K7F258 W5UIN2 R7TG96 A0A2D0R3S8 A0A2D0R5C2 F7E9S8
PDB
5AYL
E-value=9.5118e-51,
Score=509
Ontologies
GO
GO:0005623
GO:0045454
GO:0005215
GO:0055085
GO:0016021
GO:0005739
GO:0046872
GO:0051603
GO:0004222
GO:0005783
GO:0015036
GO:0034975
GO:0016671
GO:0030433
GO:0005788
GO:0015035
GO:0001933
GO:0034663
GO:0051787
GO:0051087
GO:0051117
GO:0070059
GO:0030544
GO:0001671
GO:0016853
GO:0003756
GO:0046982
GO:0006412
GO:0005737
GO:0006520
GO:0006508
GO:0008237
GO:0016020
GO:0003707
GO:0051537
Topology
Subcellular location
Endoplasmic reticulum lumen
SignalP
Position: 1 - 18,
Likelihood: 0.867973
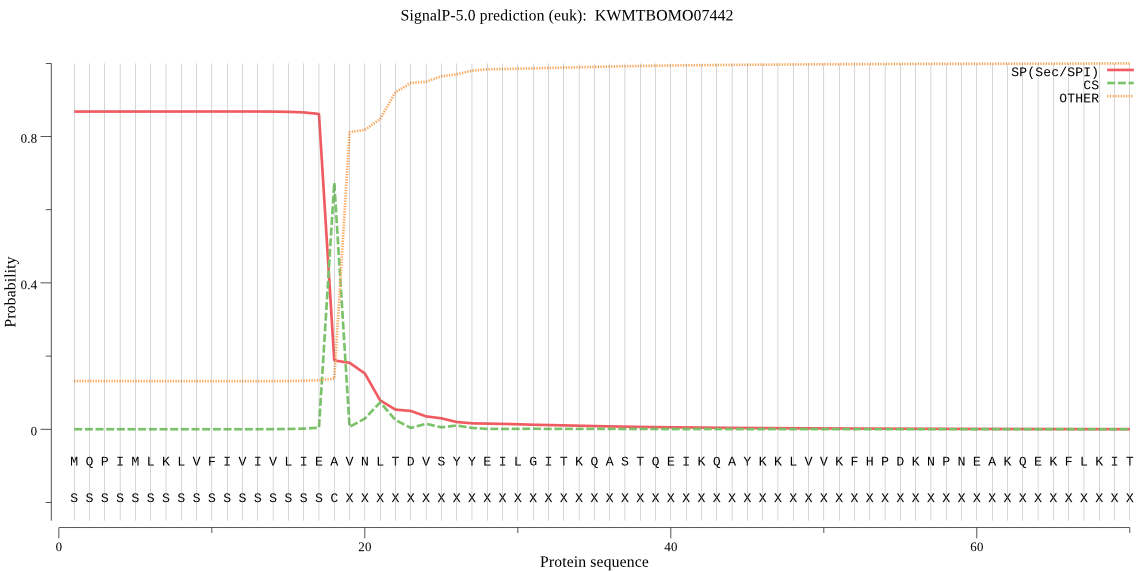
Length:
782
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.8456
Exp number, first 60 AAs:
1.82253
Total prob of N-in:
0.08387
outside
1 - 782
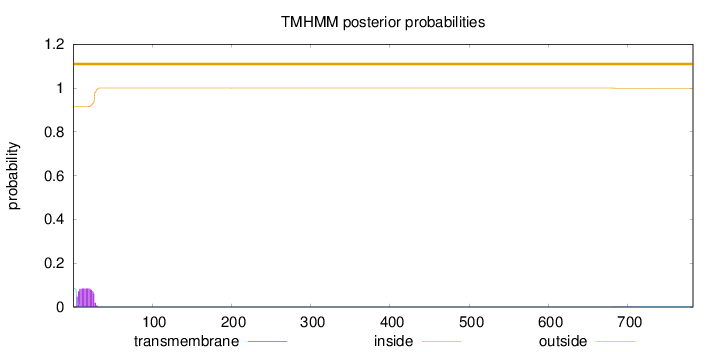
Population Genetic Test Statistics
Pi
23.899893
Theta
24.516473
Tajima's D
0.363733
CLR
0.864348
CSRT
0.47967601619919
Interpretation
Uncertain
