Gene
KWMTBOMO07415 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010351
Annotation
PREDICTED:_NADH_dehydrogenase_[ubiquinone]_1_subunit_C2_[Bombyx_mori]
Full name
NADH dehydrogenase [ubiquinone] 1 subunit C2
Location in the cell
Cytoplasmic Reliability : 1.269 Mitochondrial Reliability : 1.885
Sequence
CDS
ATGTCGCTGCATATGTCTGCCCAAGAACTCCTAAAATTAGGAGATGAAGGACGACAAAAACCTTTTCTGAACAAATATTGGCCTGAAATAATAGGAGTTACGTTCGGTATTGGCACAGGAATGTTTCTAAACTTCCAGACTAAACGGCCAGTGTTCAGCGGTATCCAAAAACATATAATTGCAACTGGAGGTTGGGTCAGCATTTTAAGTTATGTACAGCATAAACGGAATGAATATTTAGCAGAAAAGGATGCAGTATACAGGCATTATATAGAATTACACCCAGATGATTTCCCTGTTCCCGAACGTAAGAAGATTGGTGACTTGTTTGAACCTTGGATCCCAGTTCGTTAA
Protein
MSLHMSAQELLKLGDEGRQKPFLNKYWPEIIGVTFGIGTGMFLNFQTKRPVFSGIQKHIIATGGWVSILSYVQHKRNEYLAEKDAVYRHYIELHPDDFPVPERKKIGDLFEPWIPVR
Summary
Description
Accessory subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I), that is believed not to be involved in catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone.
Similarity
Belongs to the complex I NDUFC2 subunit family.
Uniprot
H9JLF0
A0A194QN68
I4DKC9
A0A194PUY6
S4NWY8
A0A0L7LFM4
+ More
A0A212FLJ7 A0A2H1W081 A0A2A4K4U1 A0A0M3QTA5 A0A034WRS1 A0A0A1WJU7 A0A1L8E192 A0A1W4UTN9 B4MZB2 B3NAR2 A0A0K8WKW3 B4I2W1 B4Q938 Q9VQM2 A0A1B0CR45 B4JAN4 B4NXE1 W8C6B2 B3N201 V5GC13 A0A1B6DXD0 A0A1Q3FJZ5 A0A0T6B269 A0A3B0JIZ0 A0A023EF40 B4LU29 U5EFI4 A0A182VYP5 Q29L28 B4GSG2 B0WA11 A0A1A9WXQ4 A0A182MRT2 D2A2K1 A0A1Y1MJW8 A0A182SHI8 A0A1Y1MJY3 A0A182XW89 Q178D3 A0A182RP24 R4WTL5 A0A1B0AYY5 A0A0K8TTF5 U4UJU1 A0A182K0S6 A0A1A9V829 N6TXN7 A0A1B0A4E0 A0A182P7R6 T1GTF6 A0A084VT43 A0A1J1HN34 A0A182N710 A0A0L0CFW7 A0A182IVQ2 A0A0A9WHA6 A0A182F977 A0A182V5K6 A0A182UAY2 A0A182LG97 A0A182XHJ6 Q7QD00 A0A182HTU7 A0A2M4C3Y0 T1E7M9 W5JRX8 A0A1B0FAB0 A0A2M4ACS9 A0A1I8N511 A0A2P8YVN3 A0A2M3Z3V2 A0A1I8P3R5 A0A336L1X5 A0A182QKW3 A0A067RFC4 A0A2S2QKE7 A0A1W4XC11 B4KEV1 A0A2H8TQT7 A0A1L8EEU8 A0A1A9XEI2 A0A224Y0K0 C4WSG2 J9KA50 A0A1B6FQZ5 A0A2S2PUN6 B7P949 A0A161N1C1 A0A0V0G4I3 A0A1B6IU48 A0A182HA83 A0A0P4VMY5 R4G4B1 A0A131Z509
A0A212FLJ7 A0A2H1W081 A0A2A4K4U1 A0A0M3QTA5 A0A034WRS1 A0A0A1WJU7 A0A1L8E192 A0A1W4UTN9 B4MZB2 B3NAR2 A0A0K8WKW3 B4I2W1 B4Q938 Q9VQM2 A0A1B0CR45 B4JAN4 B4NXE1 W8C6B2 B3N201 V5GC13 A0A1B6DXD0 A0A1Q3FJZ5 A0A0T6B269 A0A3B0JIZ0 A0A023EF40 B4LU29 U5EFI4 A0A182VYP5 Q29L28 B4GSG2 B0WA11 A0A1A9WXQ4 A0A182MRT2 D2A2K1 A0A1Y1MJW8 A0A182SHI8 A0A1Y1MJY3 A0A182XW89 Q178D3 A0A182RP24 R4WTL5 A0A1B0AYY5 A0A0K8TTF5 U4UJU1 A0A182K0S6 A0A1A9V829 N6TXN7 A0A1B0A4E0 A0A182P7R6 T1GTF6 A0A084VT43 A0A1J1HN34 A0A182N710 A0A0L0CFW7 A0A182IVQ2 A0A0A9WHA6 A0A182F977 A0A182V5K6 A0A182UAY2 A0A182LG97 A0A182XHJ6 Q7QD00 A0A182HTU7 A0A2M4C3Y0 T1E7M9 W5JRX8 A0A1B0FAB0 A0A2M4ACS9 A0A1I8N511 A0A2P8YVN3 A0A2M3Z3V2 A0A1I8P3R5 A0A336L1X5 A0A182QKW3 A0A067RFC4 A0A2S2QKE7 A0A1W4XC11 B4KEV1 A0A2H8TQT7 A0A1L8EEU8 A0A1A9XEI2 A0A224Y0K0 C4WSG2 J9KA50 A0A1B6FQZ5 A0A2S2PUN6 B7P949 A0A161N1C1 A0A0V0G4I3 A0A1B6IU48 A0A182HA83 A0A0P4VMY5 R4G4B1 A0A131Z509
Pubmed
19121390
26354079
22651552
23622113
26227816
22118469
+ More
25348373 25830018 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24495485 24945155 15632085 18362917 19820115 28004739 25244985 17510324 23691247 26369729 23537049 24438588 26108605 25401762 20966253 12364791 14747013 17210077 20920257 23761445 25315136 29403074 24845553 26483478 27129103 26830274
25348373 25830018 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24495485 24945155 15632085 18362917 19820115 28004739 25244985 17510324 23691247 26369729 23537049 24438588 26108605 25401762 20966253 12364791 14747013 17210077 20920257 23761445 25315136 29403074 24845553 26483478 27129103 26830274
EMBL
BABH01037199
BABH01037200
KQ461194
KPJ06968.1
AK401747
BAM18369.1
+ More
KQ459592 KPI96803.1 GAIX01012367 JAA80193.1 JTDY01001329 KOB74180.1 AGBW02007778 OWR54570.1 ODYU01005458 SOQ46336.1 NWSH01000162 PCG78928.1 CP012523 ALC38573.1 GAKP01001618 JAC57334.1 GBXI01015609 JAC98682.1 GFDF01001650 JAV12434.1 CH963913 EDW77385.1 CH954177 EDV57585.1 GDHF01000521 JAI51793.1 CH480820 EDW54106.1 CM000361 CM002910 EDX03577.1 KMY87824.1 AE014134 AY119127 KX532036 AAF51143.1 AAM50987.1 ANY27846.1 AJWK01024389 CH916368 EDW03842.1 CM000157 EDW87498.1 GAMC01001217 JAC05339.1 CH902667 EDV30611.1 GALX01000771 JAB67695.1 GEDC01006973 JAS30325.1 GFDL01007223 JAV27822.1 LJIG01016327 KRT80935.1 OUUW01000006 SPP82195.1 GAPW01006219 JAC07379.1 CH940649 EDW65082.1 GANO01003834 JAB56037.1 CH379061 EAL32996.2 CH479189 EDW25321.1 DS231868 EDS40744.1 AXCM01005921 KQ971338 EFA02843.1 GEZM01029622 GEZM01029621 GEZM01029619 JAV86039.1 GEZM01029620 JAV86042.1 CH477366 EAT42520.1 AK418032 BAN21247.1 JXJN01006028 GDAI01000162 JAI17441.1 KB632354 ERL93367.1 APGK01057291 KB741279 ENN71052.1 CAQQ02067388 ATLV01016167 KE525060 KFB41137.1 CVRI01000012 CRK89461.1 JRES01000438 KNC31308.1 GBHO01036813 GBRD01016461 JAG06791.1 JAG49365.1 AAAB01008859 EAA08018.3 APCN01000586 GGFJ01010872 MBW60013.1 GAMD01002892 JAA98698.1 ADMH02000272 ETN67162.1 CCAG010012870 GGFK01005239 MBW38560.1 PYGN01000334 PSN48308.1 GGFM01002448 MBW23199.1 UFQS01001498 UFQT01001020 UFQT01001498 SSX11256.1 SSX28514.1 AXCN02000322 KK852669 KDR18848.1 GGMS01009013 MBY78216.1 CH933807 EDW13001.1 GFXV01003813 GFXV01007661 MBW15618.1 MBW19466.1 GFDG01001615 JAV17184.1 GFTR01001926 JAW14500.1 AK340177 BAH70832.1 ABLF02030293 GECZ01017336 JAS52433.1 GGMR01020476 MBY33095.1 ABJB010973418 DS661587 EEC03121.1 GEMB01002299 JAS00882.1 GECL01003177 JAP02947.1 GECU01017249 JAS90457.1 JXUM01030861 JXUM01030862 JXUM01030863 KQ560899 KXJ80425.1 GDKW01000652 JAI55943.1 ACPB03003284 GAHY01000921 JAA76589.1 GEDV01002712 JAP85845.1
KQ459592 KPI96803.1 GAIX01012367 JAA80193.1 JTDY01001329 KOB74180.1 AGBW02007778 OWR54570.1 ODYU01005458 SOQ46336.1 NWSH01000162 PCG78928.1 CP012523 ALC38573.1 GAKP01001618 JAC57334.1 GBXI01015609 JAC98682.1 GFDF01001650 JAV12434.1 CH963913 EDW77385.1 CH954177 EDV57585.1 GDHF01000521 JAI51793.1 CH480820 EDW54106.1 CM000361 CM002910 EDX03577.1 KMY87824.1 AE014134 AY119127 KX532036 AAF51143.1 AAM50987.1 ANY27846.1 AJWK01024389 CH916368 EDW03842.1 CM000157 EDW87498.1 GAMC01001217 JAC05339.1 CH902667 EDV30611.1 GALX01000771 JAB67695.1 GEDC01006973 JAS30325.1 GFDL01007223 JAV27822.1 LJIG01016327 KRT80935.1 OUUW01000006 SPP82195.1 GAPW01006219 JAC07379.1 CH940649 EDW65082.1 GANO01003834 JAB56037.1 CH379061 EAL32996.2 CH479189 EDW25321.1 DS231868 EDS40744.1 AXCM01005921 KQ971338 EFA02843.1 GEZM01029622 GEZM01029621 GEZM01029619 JAV86039.1 GEZM01029620 JAV86042.1 CH477366 EAT42520.1 AK418032 BAN21247.1 JXJN01006028 GDAI01000162 JAI17441.1 KB632354 ERL93367.1 APGK01057291 KB741279 ENN71052.1 CAQQ02067388 ATLV01016167 KE525060 KFB41137.1 CVRI01000012 CRK89461.1 JRES01000438 KNC31308.1 GBHO01036813 GBRD01016461 JAG06791.1 JAG49365.1 AAAB01008859 EAA08018.3 APCN01000586 GGFJ01010872 MBW60013.1 GAMD01002892 JAA98698.1 ADMH02000272 ETN67162.1 CCAG010012870 GGFK01005239 MBW38560.1 PYGN01000334 PSN48308.1 GGFM01002448 MBW23199.1 UFQS01001498 UFQT01001020 UFQT01001498 SSX11256.1 SSX28514.1 AXCN02000322 KK852669 KDR18848.1 GGMS01009013 MBY78216.1 CH933807 EDW13001.1 GFXV01003813 GFXV01007661 MBW15618.1 MBW19466.1 GFDG01001615 JAV17184.1 GFTR01001926 JAW14500.1 AK340177 BAH70832.1 ABLF02030293 GECZ01017336 JAS52433.1 GGMR01020476 MBY33095.1 ABJB010973418 DS661587 EEC03121.1 GEMB01002299 JAS00882.1 GECL01003177 JAP02947.1 GECU01017249 JAS90457.1 JXUM01030861 JXUM01030862 JXUM01030863 KQ560899 KXJ80425.1 GDKW01000652 JAI55943.1 ACPB03003284 GAHY01000921 JAA76589.1 GEDV01002712 JAP85845.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000037510
UP000007151
UP000218220
+ More
UP000092553 UP000192221 UP000007798 UP000008711 UP000001292 UP000000304 UP000000803 UP000092461 UP000001070 UP000002282 UP000007801 UP000268350 UP000008792 UP000075920 UP000001819 UP000008744 UP000002320 UP000091820 UP000075883 UP000007266 UP000075901 UP000076408 UP000008820 UP000075900 UP000092460 UP000030742 UP000075881 UP000078200 UP000019118 UP000092445 UP000075885 UP000015102 UP000030765 UP000183832 UP000075884 UP000037069 UP000075880 UP000069272 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000000673 UP000092444 UP000095301 UP000245037 UP000095300 UP000075886 UP000027135 UP000192223 UP000009192 UP000092443 UP000007819 UP000001555 UP000069940 UP000249989 UP000015103
UP000092553 UP000192221 UP000007798 UP000008711 UP000001292 UP000000304 UP000000803 UP000092461 UP000001070 UP000002282 UP000007801 UP000268350 UP000008792 UP000075920 UP000001819 UP000008744 UP000002320 UP000091820 UP000075883 UP000007266 UP000075901 UP000076408 UP000008820 UP000075900 UP000092460 UP000030742 UP000075881 UP000078200 UP000019118 UP000092445 UP000075885 UP000015102 UP000030765 UP000183832 UP000075884 UP000037069 UP000075880 UP000069272 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000000673 UP000092444 UP000095301 UP000245037 UP000095300 UP000075886 UP000027135 UP000192223 UP000009192 UP000092443 UP000007819 UP000001555 UP000069940 UP000249989 UP000015103
Pfam
PF06374 NDUF_C2
Interpro
IPR009423
NADH-UbQ_OxRdtase_b14.5b_su
ProteinModelPortal
H9JLF0
A0A194QN68
I4DKC9
A0A194PUY6
S4NWY8
A0A0L7LFM4
+ More
A0A212FLJ7 A0A2H1W081 A0A2A4K4U1 A0A0M3QTA5 A0A034WRS1 A0A0A1WJU7 A0A1L8E192 A0A1W4UTN9 B4MZB2 B3NAR2 A0A0K8WKW3 B4I2W1 B4Q938 Q9VQM2 A0A1B0CR45 B4JAN4 B4NXE1 W8C6B2 B3N201 V5GC13 A0A1B6DXD0 A0A1Q3FJZ5 A0A0T6B269 A0A3B0JIZ0 A0A023EF40 B4LU29 U5EFI4 A0A182VYP5 Q29L28 B4GSG2 B0WA11 A0A1A9WXQ4 A0A182MRT2 D2A2K1 A0A1Y1MJW8 A0A182SHI8 A0A1Y1MJY3 A0A182XW89 Q178D3 A0A182RP24 R4WTL5 A0A1B0AYY5 A0A0K8TTF5 U4UJU1 A0A182K0S6 A0A1A9V829 N6TXN7 A0A1B0A4E0 A0A182P7R6 T1GTF6 A0A084VT43 A0A1J1HN34 A0A182N710 A0A0L0CFW7 A0A182IVQ2 A0A0A9WHA6 A0A182F977 A0A182V5K6 A0A182UAY2 A0A182LG97 A0A182XHJ6 Q7QD00 A0A182HTU7 A0A2M4C3Y0 T1E7M9 W5JRX8 A0A1B0FAB0 A0A2M4ACS9 A0A1I8N511 A0A2P8YVN3 A0A2M3Z3V2 A0A1I8P3R5 A0A336L1X5 A0A182QKW3 A0A067RFC4 A0A2S2QKE7 A0A1W4XC11 B4KEV1 A0A2H8TQT7 A0A1L8EEU8 A0A1A9XEI2 A0A224Y0K0 C4WSG2 J9KA50 A0A1B6FQZ5 A0A2S2PUN6 B7P949 A0A161N1C1 A0A0V0G4I3 A0A1B6IU48 A0A182HA83 A0A0P4VMY5 R4G4B1 A0A131Z509
A0A212FLJ7 A0A2H1W081 A0A2A4K4U1 A0A0M3QTA5 A0A034WRS1 A0A0A1WJU7 A0A1L8E192 A0A1W4UTN9 B4MZB2 B3NAR2 A0A0K8WKW3 B4I2W1 B4Q938 Q9VQM2 A0A1B0CR45 B4JAN4 B4NXE1 W8C6B2 B3N201 V5GC13 A0A1B6DXD0 A0A1Q3FJZ5 A0A0T6B269 A0A3B0JIZ0 A0A023EF40 B4LU29 U5EFI4 A0A182VYP5 Q29L28 B4GSG2 B0WA11 A0A1A9WXQ4 A0A182MRT2 D2A2K1 A0A1Y1MJW8 A0A182SHI8 A0A1Y1MJY3 A0A182XW89 Q178D3 A0A182RP24 R4WTL5 A0A1B0AYY5 A0A0K8TTF5 U4UJU1 A0A182K0S6 A0A1A9V829 N6TXN7 A0A1B0A4E0 A0A182P7R6 T1GTF6 A0A084VT43 A0A1J1HN34 A0A182N710 A0A0L0CFW7 A0A182IVQ2 A0A0A9WHA6 A0A182F977 A0A182V5K6 A0A182UAY2 A0A182LG97 A0A182XHJ6 Q7QD00 A0A182HTU7 A0A2M4C3Y0 T1E7M9 W5JRX8 A0A1B0FAB0 A0A2M4ACS9 A0A1I8N511 A0A2P8YVN3 A0A2M3Z3V2 A0A1I8P3R5 A0A336L1X5 A0A182QKW3 A0A067RFC4 A0A2S2QKE7 A0A1W4XC11 B4KEV1 A0A2H8TQT7 A0A1L8EEU8 A0A1A9XEI2 A0A224Y0K0 C4WSG2 J9KA50 A0A1B6FQZ5 A0A2S2PUN6 B7P949 A0A161N1C1 A0A0V0G4I3 A0A1B6IU48 A0A182HA83 A0A0P4VMY5 R4G4B1 A0A131Z509
PDB
6G72
E-value=1.02899e-06,
Score=119
Ontologies
PATHWAY
GO
PANTHER
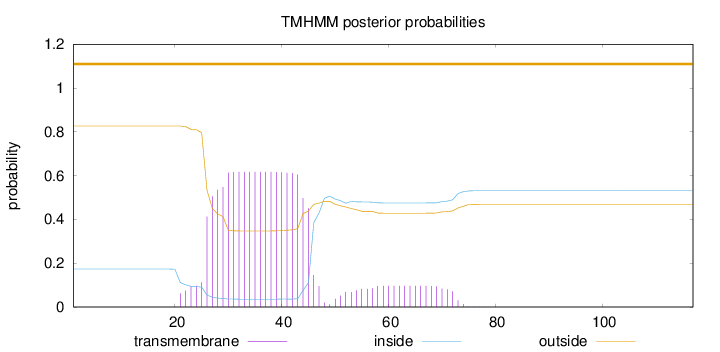
Topology
Subcellular location
Mitochondrion inner membrane
Length:
117
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
14.26198
Exp number, first 60 AAs:
13.11341
Total prob of N-in:
0.17308
POSSIBLE N-term signal
sequence
outside
1 - 117
Population Genetic Test Statistics
Pi
36.847592
Theta
29.738926
Tajima's D
0.628524
CLR
0.266218
CSRT
0.55657217139143
Interpretation
Uncertain
