Gene
KWMTBOMO07413
Pre Gene Modal
BGIBMGA010350
Annotation
hypothetical_protein_KGM_04267_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 4.351
Sequence
CDS
ATGGCTGATATTCTTGATATATTAGACATAGAACAACCGGCAACAGAGATCAGCAGGGACAGTATTATACATGGCGATAAGAACAAAAAGAAATATGTTACCGCTAAAGCTGTCAAGAGGCCCGAGGGAATGCATAGGGAGGTTTTCGCATTACTTTATAATGACAATAAGGAATTACCTCCTTTACTGCCTACTGATACGGGAAAAGCATACAAACAGACAAAAGCCAGGCTTGGCATGAGGAAAGTCAGAAAATGGGTATGGGCACCATTTACTAATCCTGCACGTAAAGACAATGCAGTTTTCCACCACTGGAAGAGGGCATCTGATGAAGCTAAAGAATACCCCTTTGCACAATTCAATAAGCAAGTGTCAATACCATCTTATTCCGAGTCCGAATATAACCAATACTTAAAATCTGAAGATTGGAGTCAAGCAGAAACTGATCATCTGATGGATTTATGCCAACGTTTTGATTTGCGATTTATTGTAATACATGATCGATGGGACAGGGCTGCATTCAGAGATCGAAGTGTAGAAGATTTGAAGGAACGTTATTATAATATCTGCTCTATTCTGAGTAAGGTTAAGACTAATCCCTGGTCAAACACAGTTACAATGGTGAATGGCGAAAAACGCGTCTATCATTACGATGCTGAACATGAACGAAAGAGAAAGGAACAACTGCGAAGACTATTTGATCGCACTCAAGAACAGATCGATGAAGAGCAAATGCTGCTAGCTGAACTGAAAAAGATCGAAGCCAGGAAACGGGAAAGGGAAAGGAAAACTCAAGATTTACAGAAACTTATATCAAGAGCAGACAGCGGCACTATTACCCCATCAACTCCGACAAACCATAACACAGAGCCCCCAGGAACACCTTCAAGCATGGCTGCCAACATGAGGAGGCATGATCGGAAATTACATAAGAAGAAGATAACCACACAGCAAAGACCCATGCGTGCTGTGGAAAATGTGACAGTGGAGTGGTCAGGAATAAAGTTTCCGGAGGCTCGAGGAACTGGTGTATGGCTACGCTCTCAACGCATGAAACTGCCTCCTGGAGTAGGACAACGTAAAACGAAAGCTATTGAACAAGAGCTTCGACTAATGAACATAGATATAGCACCAACTCCAACTGAACCTATCTGCAAACATTTCAATGAGCTCCGCTCTGATCTGGCTTTGTCCATAGATCTGAAAAATGCTTTAGCATCCTGTGAGTTTGAATTGCAAGCTTTGCGCCACCAATATGAAGCTTTAAACCCGGGAAAGACCCTTACGATTCCAGCATCAATATGCAACACAAGTGTGGACAGTGACTCGAAACCAGTTGGTGAAATAATTGATGTGGTTGGTTCTCCTGGTGCAATGAATACAACCATATAG
Protein
MADILDILDIEQPATEISRDSIIHGDKNKKKYVTAKAVKRPEGMHREVFALLYNDNKELPPLLPTDTGKAYKQTKARLGMRKVRKWVWAPFTNPARKDNAVFHHWKRASDEAKEYPFAQFNKQVSIPSYSESEYNQYLKSEDWSQAETDHLMDLCQRFDLRFIVIHDRWDRAAFRDRSVEDLKERYYNICSILSKVKTNPWSNTVTMVNGEKRVYHYDAEHERKRKEQLRRLFDRTQEQIDEEQMLLAELKKIEARKRERERKTQDLQKLISRADSGTITPSTPTNHNTEPPGTPSSMAANMRRHDRKLHKKKITTQQRPMRAVENVTVEWSGIKFPEARGTGVWLRSQRMKLPPGVGQRKTKAIEQELRLMNIDIAPTPTEPICKHFNELRSDLALSIDLKNALASCEFELQALRHQYEALNPGKTLTIPASICNTSVDSDSKPVGEIIDVVGSPGAMNTTI
Summary
Uniprot
A0A2A4K4H0
A0A212ETZ3
H9JLE9
A0A1E1WNX5
A0A194PTR0
A0A194QTU8
+ More
A0A3S2PAS4 A0A2H1W7L6 A0A1W4WXS5 A0A067RIT0 A0A1W4X8C4 A0A2J7PVR4 A0A0A9Y3B2 A0A1Y1MBL1 E2BII3 N6TB19 A0A0V0G9V8 A0A069DU36 A0A023EXH5 U4UHB0 A0A0P4W4M4 A0A1B6C5W3 E2A0I5 A0A088A2P7 D6WR58 A0A2A3EH17 A0A1B6KEB7 A0A0L7RBE0 A0A2J7PVS4 A0A1B0CH62 T1HN61 A0A0C9QLH4 A0A336MQ36 F4WZ13 A0A158NDL3 A0A1L8DX30 A0A232EZ72 A0A067ZYD1 A0A195CN04 A0A182G7U7 A0A182PQK6 A0A182U9G3 Q7QJA8 W5JVB8 A0A182VBL4 A0A182KR66 A0A182I354 A0A1L8DWV0 A0A182NI11 A0A151XEJ1 A0A195F505 A0A026WHJ1 A0A182Q9K1 Q16TX1 E9GVW3 A0A182JR06 A0A195DA09 A0A182XYQ6 A0A0L0C360 B0XFR8 A0A182S7X4 A0A2R5LJE1 A0A1Q3F8V9 A0A0P5UW99 A0A0N7ZTW8 A0A182G5H0 A0A154PNR1 A0A0M9A137 A0A084WN28 U5ET58 A0A2L2YAR3 A0A0K8TLB2 A0A0P5N6Z3 A0A0P5USK0 A0A0A1WID9 A0A1I8MEK3 A0A0P5N171 T1P836 A0A0P5BAM9 A0A0P4XXD3 A0A0K8WG49 A0A034WAY7 W8BPL6 A0A1I8PSB2 A0A195BQ82 A0A0J7KIQ1 A0A1A9YE44 A0A1B0BC14 B3NJW5 B3MIT8 V5H394 A0A131Y0S4 A0A1B0F9E1 B4PA34 B4QF37 B4HQM9 B4ME98 A0A1A9WWJ7
A0A3S2PAS4 A0A2H1W7L6 A0A1W4WXS5 A0A067RIT0 A0A1W4X8C4 A0A2J7PVR4 A0A0A9Y3B2 A0A1Y1MBL1 E2BII3 N6TB19 A0A0V0G9V8 A0A069DU36 A0A023EXH5 U4UHB0 A0A0P4W4M4 A0A1B6C5W3 E2A0I5 A0A088A2P7 D6WR58 A0A2A3EH17 A0A1B6KEB7 A0A0L7RBE0 A0A2J7PVS4 A0A1B0CH62 T1HN61 A0A0C9QLH4 A0A336MQ36 F4WZ13 A0A158NDL3 A0A1L8DX30 A0A232EZ72 A0A067ZYD1 A0A195CN04 A0A182G7U7 A0A182PQK6 A0A182U9G3 Q7QJA8 W5JVB8 A0A182VBL4 A0A182KR66 A0A182I354 A0A1L8DWV0 A0A182NI11 A0A151XEJ1 A0A195F505 A0A026WHJ1 A0A182Q9K1 Q16TX1 E9GVW3 A0A182JR06 A0A195DA09 A0A182XYQ6 A0A0L0C360 B0XFR8 A0A182S7X4 A0A2R5LJE1 A0A1Q3F8V9 A0A0P5UW99 A0A0N7ZTW8 A0A182G5H0 A0A154PNR1 A0A0M9A137 A0A084WN28 U5ET58 A0A2L2YAR3 A0A0K8TLB2 A0A0P5N6Z3 A0A0P5USK0 A0A0A1WID9 A0A1I8MEK3 A0A0P5N171 T1P836 A0A0P5BAM9 A0A0P4XXD3 A0A0K8WG49 A0A034WAY7 W8BPL6 A0A1I8PSB2 A0A195BQ82 A0A0J7KIQ1 A0A1A9YE44 A0A1B0BC14 B3NJW5 B3MIT8 V5H394 A0A131Y0S4 A0A1B0F9E1 B4PA34 B4QF37 B4HQM9 B4ME98 A0A1A9WWJ7
Pubmed
22118469
19121390
26354079
24845553
25401762
26823975
+ More
28004739 20798317 23537049 26334808 25474469 27129103 18362917 19820115 21719571 21347285 28648823 26483478 12364791 14747013 17210077 20920257 23761445 20966253 24508170 30249741 17510324 21292972 25244985 26108605 24438588 26561354 26369729 25830018 25315136 25348373 24495485 17994087 25765539 17550304 22936249
28004739 20798317 23537049 26334808 25474469 27129103 18362917 19820115 21719571 21347285 28648823 26483478 12364791 14747013 17210077 20920257 23761445 20966253 24508170 30249741 17510324 21292972 25244985 26108605 24438588 26561354 26369729 25830018 25315136 25348373 24495485 17994087 25765539 17550304 22936249
EMBL
NWSH01000162
PCG78926.1
AGBW02012509
OWR44963.1
BABH01037198
GDQN01002362
+ More
JAT88692.1 KQ459592 KPI96801.1 KQ461194 KPJ06966.1 RSAL01000139 RVE46145.1 ODYU01006866 SOQ49095.1 KK852607 KDR20356.1 NEVH01020943 PNF20428.1 GBHO01016053 GBHO01016052 GBRD01011039 GBRD01011038 GDHC01011286 JAG27551.1 JAG27552.1 JAG54785.1 JAQ07343.1 GEZM01036107 JAV82971.1 GL448512 EFN84493.1 APGK01036914 KB740945 ENN77469.1 GECL01001954 JAP04170.1 GBGD01001464 JAC87425.1 GBBI01005066 JAC13646.1 KB632197 ERL89986.1 GDKW01000298 JAI56297.1 GEDC01028504 GEDC01024031 GEDC01017891 GEDC01014722 JAS08794.1 JAS13267.1 JAS19407.1 JAS22576.1 GL435626 EFN72888.1 KQ971354 EFA07023.2 KZ288252 PBC31027.1 GEBQ01030177 JAT09800.1 KQ414617 KOC68135.1 PNF20429.1 AJWK01012066 ACPB03002860 GBYB01004389 GBYB01004776 JAG74156.1 JAG74543.1 UFQT01001519 SSX30893.1 GL888465 EGI60516.1 ADTU01012715 GFDF01003115 JAV10969.1 NNAY01001563 OXU23587.1 KF294267 AHZ08395.1 KQ977600 KYN01469.1 JXUM01000599 KQ560108 KXJ84491.1 AAAB01008807 EAA04266.4 ADMH02000243 ETN67268.1 APCN01000197 GFDF01003143 JAV10941.1 KQ982254 KYQ58767.1 KQ981820 KYN35159.1 KK107200 QOIP01000002 EZA55500.1 RLU25322.1 AXCN02000387 CH477638 EAT37953.1 GL732569 EFX76249.1 KQ981082 KYN09697.1 JRES01000956 KNC26765.1 DS232955 EDS26997.1 GGLE01005495 MBY09621.1 GFDL01011059 JAV23986.1 GDIP01109245 JAL94469.1 GDIP01212993 LRGB01003325 JAJ10409.1 KZS03193.1 JXUM01043698 KQ561372 KXJ78813.1 KQ434977 KZC12860.1 KQ435793 KOX74226.1 ATLV01024557 KE525352 KFB51622.1 GANO01002918 JAB56953.1 IAAA01018913 IAAA01018914 LAA04450.1 GDAI01002494 JAI15109.1 GDIQ01148778 JAL02948.1 GDIP01109244 JAL94470.1 GBXI01016129 JAC98162.1 GDIQ01159926 JAK91799.1 KA644714 AFP59343.1 GDIP01187142 JAJ36260.1 GDIP01236431 JAI86970.1 GDHF01002290 JAI50024.1 GAKP01007984 JAC50968.1 GAMC01005668 JAC00888.1 KQ976424 KYM88298.1 LBMM01007007 KMQ90124.1 JXJN01011768 CH954179 EDV55340.1 CH902619 EDV35998.1 GANP01007023 JAB77445.1 GEFM01002937 JAP72859.1 CCAG010012936 CM000158 EDW91365.1 CM000362 CM002911 EDX07948.1 KMY95311.1 CH480816 EDW48732.1 CH940662 EDW58863.1
JAT88692.1 KQ459592 KPI96801.1 KQ461194 KPJ06966.1 RSAL01000139 RVE46145.1 ODYU01006866 SOQ49095.1 KK852607 KDR20356.1 NEVH01020943 PNF20428.1 GBHO01016053 GBHO01016052 GBRD01011039 GBRD01011038 GDHC01011286 JAG27551.1 JAG27552.1 JAG54785.1 JAQ07343.1 GEZM01036107 JAV82971.1 GL448512 EFN84493.1 APGK01036914 KB740945 ENN77469.1 GECL01001954 JAP04170.1 GBGD01001464 JAC87425.1 GBBI01005066 JAC13646.1 KB632197 ERL89986.1 GDKW01000298 JAI56297.1 GEDC01028504 GEDC01024031 GEDC01017891 GEDC01014722 JAS08794.1 JAS13267.1 JAS19407.1 JAS22576.1 GL435626 EFN72888.1 KQ971354 EFA07023.2 KZ288252 PBC31027.1 GEBQ01030177 JAT09800.1 KQ414617 KOC68135.1 PNF20429.1 AJWK01012066 ACPB03002860 GBYB01004389 GBYB01004776 JAG74156.1 JAG74543.1 UFQT01001519 SSX30893.1 GL888465 EGI60516.1 ADTU01012715 GFDF01003115 JAV10969.1 NNAY01001563 OXU23587.1 KF294267 AHZ08395.1 KQ977600 KYN01469.1 JXUM01000599 KQ560108 KXJ84491.1 AAAB01008807 EAA04266.4 ADMH02000243 ETN67268.1 APCN01000197 GFDF01003143 JAV10941.1 KQ982254 KYQ58767.1 KQ981820 KYN35159.1 KK107200 QOIP01000002 EZA55500.1 RLU25322.1 AXCN02000387 CH477638 EAT37953.1 GL732569 EFX76249.1 KQ981082 KYN09697.1 JRES01000956 KNC26765.1 DS232955 EDS26997.1 GGLE01005495 MBY09621.1 GFDL01011059 JAV23986.1 GDIP01109245 JAL94469.1 GDIP01212993 LRGB01003325 JAJ10409.1 KZS03193.1 JXUM01043698 KQ561372 KXJ78813.1 KQ434977 KZC12860.1 KQ435793 KOX74226.1 ATLV01024557 KE525352 KFB51622.1 GANO01002918 JAB56953.1 IAAA01018913 IAAA01018914 LAA04450.1 GDAI01002494 JAI15109.1 GDIQ01148778 JAL02948.1 GDIP01109244 JAL94470.1 GBXI01016129 JAC98162.1 GDIQ01159926 JAK91799.1 KA644714 AFP59343.1 GDIP01187142 JAJ36260.1 GDIP01236431 JAI86970.1 GDHF01002290 JAI50024.1 GAKP01007984 JAC50968.1 GAMC01005668 JAC00888.1 KQ976424 KYM88298.1 LBMM01007007 KMQ90124.1 JXJN01011768 CH954179 EDV55340.1 CH902619 EDV35998.1 GANP01007023 JAB77445.1 GEFM01002937 JAP72859.1 CCAG010012936 CM000158 EDW91365.1 CM000362 CM002911 EDX07948.1 KMY95311.1 CH480816 EDW48732.1 CH940662 EDW58863.1
Proteomes
UP000218220
UP000007151
UP000005204
UP000053268
UP000053240
UP000283053
+ More
UP000192223 UP000027135 UP000235965 UP000008237 UP000019118 UP000030742 UP000000311 UP000005203 UP000007266 UP000242457 UP000053825 UP000092461 UP000015103 UP000007755 UP000005205 UP000215335 UP000078542 UP000069940 UP000249989 UP000075885 UP000075902 UP000007062 UP000000673 UP000075903 UP000075882 UP000075840 UP000075884 UP000075809 UP000078541 UP000053097 UP000279307 UP000075886 UP000008820 UP000000305 UP000075881 UP000078492 UP000076408 UP000037069 UP000002320 UP000075901 UP000076858 UP000076502 UP000053105 UP000030765 UP000095301 UP000095300 UP000078540 UP000036403 UP000092443 UP000092460 UP000008711 UP000007801 UP000092444 UP000002282 UP000000304 UP000001292 UP000008792 UP000091820
UP000192223 UP000027135 UP000235965 UP000008237 UP000019118 UP000030742 UP000000311 UP000005203 UP000007266 UP000242457 UP000053825 UP000092461 UP000015103 UP000007755 UP000005205 UP000215335 UP000078542 UP000069940 UP000249989 UP000075885 UP000075902 UP000007062 UP000000673 UP000075903 UP000075882 UP000075840 UP000075884 UP000075809 UP000078541 UP000053097 UP000279307 UP000075886 UP000008820 UP000000305 UP000075881 UP000078492 UP000076408 UP000037069 UP000002320 UP000075901 UP000076858 UP000076502 UP000053105 UP000030765 UP000095301 UP000095300 UP000078540 UP000036403 UP000092443 UP000092460 UP000008711 UP000007801 UP000092444 UP000002282 UP000000304 UP000001292 UP000008792 UP000091820
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A2A4K4H0
A0A212ETZ3
H9JLE9
A0A1E1WNX5
A0A194PTR0
A0A194QTU8
+ More
A0A3S2PAS4 A0A2H1W7L6 A0A1W4WXS5 A0A067RIT0 A0A1W4X8C4 A0A2J7PVR4 A0A0A9Y3B2 A0A1Y1MBL1 E2BII3 N6TB19 A0A0V0G9V8 A0A069DU36 A0A023EXH5 U4UHB0 A0A0P4W4M4 A0A1B6C5W3 E2A0I5 A0A088A2P7 D6WR58 A0A2A3EH17 A0A1B6KEB7 A0A0L7RBE0 A0A2J7PVS4 A0A1B0CH62 T1HN61 A0A0C9QLH4 A0A336MQ36 F4WZ13 A0A158NDL3 A0A1L8DX30 A0A232EZ72 A0A067ZYD1 A0A195CN04 A0A182G7U7 A0A182PQK6 A0A182U9G3 Q7QJA8 W5JVB8 A0A182VBL4 A0A182KR66 A0A182I354 A0A1L8DWV0 A0A182NI11 A0A151XEJ1 A0A195F505 A0A026WHJ1 A0A182Q9K1 Q16TX1 E9GVW3 A0A182JR06 A0A195DA09 A0A182XYQ6 A0A0L0C360 B0XFR8 A0A182S7X4 A0A2R5LJE1 A0A1Q3F8V9 A0A0P5UW99 A0A0N7ZTW8 A0A182G5H0 A0A154PNR1 A0A0M9A137 A0A084WN28 U5ET58 A0A2L2YAR3 A0A0K8TLB2 A0A0P5N6Z3 A0A0P5USK0 A0A0A1WID9 A0A1I8MEK3 A0A0P5N171 T1P836 A0A0P5BAM9 A0A0P4XXD3 A0A0K8WG49 A0A034WAY7 W8BPL6 A0A1I8PSB2 A0A195BQ82 A0A0J7KIQ1 A0A1A9YE44 A0A1B0BC14 B3NJW5 B3MIT8 V5H394 A0A131Y0S4 A0A1B0F9E1 B4PA34 B4QF37 B4HQM9 B4ME98 A0A1A9WWJ7
A0A3S2PAS4 A0A2H1W7L6 A0A1W4WXS5 A0A067RIT0 A0A1W4X8C4 A0A2J7PVR4 A0A0A9Y3B2 A0A1Y1MBL1 E2BII3 N6TB19 A0A0V0G9V8 A0A069DU36 A0A023EXH5 U4UHB0 A0A0P4W4M4 A0A1B6C5W3 E2A0I5 A0A088A2P7 D6WR58 A0A2A3EH17 A0A1B6KEB7 A0A0L7RBE0 A0A2J7PVS4 A0A1B0CH62 T1HN61 A0A0C9QLH4 A0A336MQ36 F4WZ13 A0A158NDL3 A0A1L8DX30 A0A232EZ72 A0A067ZYD1 A0A195CN04 A0A182G7U7 A0A182PQK6 A0A182U9G3 Q7QJA8 W5JVB8 A0A182VBL4 A0A182KR66 A0A182I354 A0A1L8DWV0 A0A182NI11 A0A151XEJ1 A0A195F505 A0A026WHJ1 A0A182Q9K1 Q16TX1 E9GVW3 A0A182JR06 A0A195DA09 A0A182XYQ6 A0A0L0C360 B0XFR8 A0A182S7X4 A0A2R5LJE1 A0A1Q3F8V9 A0A0P5UW99 A0A0N7ZTW8 A0A182G5H0 A0A154PNR1 A0A0M9A137 A0A084WN28 U5ET58 A0A2L2YAR3 A0A0K8TLB2 A0A0P5N6Z3 A0A0P5USK0 A0A0A1WID9 A0A1I8MEK3 A0A0P5N171 T1P836 A0A0P5BAM9 A0A0P4XXD3 A0A0K8WG49 A0A034WAY7 W8BPL6 A0A1I8PSB2 A0A195BQ82 A0A0J7KIQ1 A0A1A9YE44 A0A1B0BC14 B3NJW5 B3MIT8 V5H394 A0A131Y0S4 A0A1B0F9E1 B4PA34 B4QF37 B4HQM9 B4ME98 A0A1A9WWJ7
PDB
4IEJ
E-value=4.77988e-28,
Score=311
Ontologies
GO
PANTHER
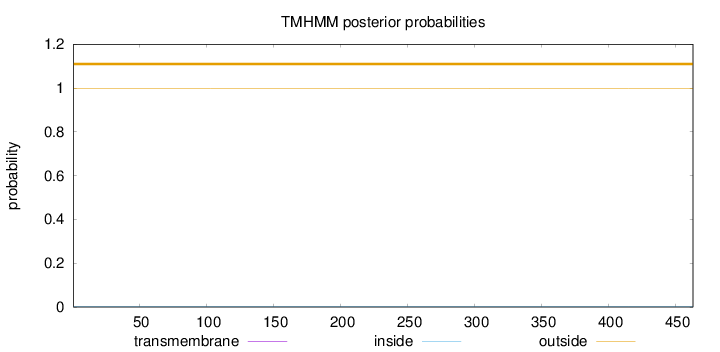
Topology
Length:
463
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00128
outside
1 - 463
Population Genetic Test Statistics
Pi
33.113207
Theta
27.735646
Tajima's D
0.562518
CLR
0.549783
CSRT
0.546022698865057
Interpretation
Uncertain
