Gene
KWMTBOMO07412 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010349
Annotation
PREDICTED:_serine_carboxypeptidase-like_51_[Amyelois_transitella]
Full name
Carboxypeptidase
Location in the cell
PlasmaMembrane Reliability : 3.243
Sequence
CDS
ATGTCGAGGAAGACTAACATAATAATAGTGTCAGTTATATTAGGTTTTGTGGCAGCAGCTGTGATTGGATTTCTAATATGGTGGCTGGTAACTGATACAGAGGAATTTACCATAATCAAACTCGAAGGTGAAGGCATTGGTGATATTGAATACACAGCAGCATTTACGAGAATAAGGGGGAGCGGCGATATATTTTGGTGGTTCTATCCCACTCTCCCTGCTGTCAAAATCATAAAACCTATTATATTGTGGGTGGATGGCGTCACTGGTGTACCGCCTAGTCTACTGGCAAATTTCGGCATGTTCGGACCGTACGACTTCAACTTGAACCAACGGAACGATTCTTGGATTAATAACTACAATTTACTATTCGTGGATGCACCGCTTGGTACAGGATTTAGTACAGCCCAGAGCGACAGTCAGATTCCAGGGAACCTTGACGCGACTACTGACCATCTGTTGTTTACCTTGGAATCATTTTACCGTCTTCATGAAGACTACGCAGATACCCCACTCTATATTTTCGGTGAAGGGCATGGTGCGCAAATAGCATTGGCTCTCGCCAGGAGGCTTGCCTCAACCGATCCACCTGGCAGGTTCCAGCACAATTTGAAAGGAGTCGTACTCGGTAACGGAATTGTTTCTCCTGCTCTCGCCTTGACCAAACTGGGATTTTATTTAGAAGAACTTGGTTACATCGATGCTAACGGGAGGGCGGCCATTGAGACACTTTCAGAAGACGTGAGCAGCTCGGTTAGTGGCGGTCAACTAAGAGATGCTTTTGACAAATTTATTACACTTGGAGATTTCGTGAATGAAAAAGCAGGCGCTGTCGCTGTTAACTTGGGGCATATAGTCGATAAATTGACACGGGAAGCGTCAAGAGATTATTTCGGTCAAAGACAATACATGCAGGAAAAATTCGGTTTAGACGCCTTTTCATTCATGCAGGAAACCGTGGCTCCTGCTTTGGGTATACCATCAGACGTTTCGTTTGACTACAACAGAGCTGCTGTCGTTGATGCATTCAGAAACAATTTTATGGCACCAGCCACACCTCACGTTGAATTCCTTCTTAGAAACACGGACTTGAAGGTGGTAATTTATAATGGAAATCTCGATGCCGTTTCCAATACTCCAGGTCAACTAGAGTGGGTAGATAATTTACAATGGCCAGGTCAGGAAGAATTCAAAACTACTCCTCGACGAACTTTAGTAATTAACAGATTAGTAGAAGGCTATTTCAGGGAAACTCCGAGATTGTCATTCTATTGGATGAACGCAGCTGGTCAATCGGTTCCACTCGACAGTCCACTAGCAATGCGACGGATTTTGGACAGGATAACACAACCCAAATGA
Protein
MSRKTNIIIVSVILGFVAAAVIGFLIWWLVTDTEEFTIIKLEGEGIGDIEYTAAFTRIRGSGDIFWWFYPTLPAVKIIKPIILWVDGVTGVPPSLLANFGMFGPYDFNLNQRNDSWINNYNLLFVDAPLGTGFSTAQSDSQIPGNLDATTDHLLFTLESFYRLHEDYADTPLYIFGEGHGAQIALALARRLASTDPPGRFQHNLKGVVLGNGIVSPALALTKLGFYLEELGYIDANGRAAIETLSEDVSSSVSGGQLRDAFDKFITLGDFVNEKAGAVAVNLGHIVDKLTREASRDYFGQRQYMQEKFGLDAFSFMQETVAPALGIPSDVSFDYNRAAVVDAFRNNFMAPATPHVEFLLRNTDLKVVIYNGNLDAVSNTPGQLEWVDNLQWPGQEEFKTTPRRTLVINRLVEGYFRETPRLSFYWMNAAGQSVPLDSPLAMRRILDRITQPK
Summary
Similarity
Belongs to the peptidase S10 family.
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Feature
chain Carboxypeptidase
Uniprot
H9JLE8
A0A1E1WNG9
A0A2A4JTZ5
A0A2A4K3U2
A0A1E1WQ57
A0A212ESC2
+ More
A0A2H1W9D8 A0A194PVL1 A0A194QNN2 D6W900 D6W8Z8 H9JLE7 D6W901 D6W8Z9 A0A182GA02 B0W744 A0A2A4JS98 A0A212ESD4 A0A2A4K4T6 A0A194QPS0 K7IP63 Q173P0 A0A158P0S0 E0VVV2 A0A194Q057 F4WUY1 A0A151WVZ7 K7J676 A0A1W4XBY5 A0A1B6CJV6 E2BXB0 A0A195FX59 A0A1E1WIS7 K7IV24 A0A2J7Q9N9 A0A232FA17 A0A1I8NA52 A0A2A3ENY1 A0A0C9R901 A0A1W4X145 B9W4M6 A0A1I8M708 A0A3L8E0Z4 A0A3G5BIH1 A0A310SIQ6 A0A1Y9J147 Q7PMZ0 A0A182YDC7 W5JIV8 A0A1E1W9K3 A0A182V9C7 A0A023GNA5 A0A1W7RAV1 A0A336L455 A0A224Z6E4 A0A182P9E3 K7IP64 A0A182RFB8 A0A0C9PYP4 J3JW05 A0A1I8N597 A0A131YXB8 A0A026VTG8 A0A0L7QX14 A0A182HNM1 A0A195AUG0 A0A1A9WW87 A0A182JYW3 A0A084WBG2 A0A1I8PQA5 A0A182TR84 W8AS88 A0A088A9Q0 A0A1Y1MRG9 A0A1I8M8P6 A0A154NZ13 A0A293MTR0 B4LGK4 A0A2I9LNU5 A0A067RRG0 A0A0A1WN20 A0A0K8W077 A0A1A9VMJ8 A0A1B0FRI3 A0A1A9Y3F6 T1J5M9 A0A3B0K462 A0A0P4WJ86 A0A1D1UPI1 A0A1B0AEJ3 N6UG27 A0A182MXX8 B4H4A7 Q29DW4 A0A0M5J3G2 A0A1B0ASD3 D6WQS3 B3ML05 A0A034WDZ5
A0A2H1W9D8 A0A194PVL1 A0A194QNN2 D6W900 D6W8Z8 H9JLE7 D6W901 D6W8Z9 A0A182GA02 B0W744 A0A2A4JS98 A0A212ESD4 A0A2A4K4T6 A0A194QPS0 K7IP63 Q173P0 A0A158P0S0 E0VVV2 A0A194Q057 F4WUY1 A0A151WVZ7 K7J676 A0A1W4XBY5 A0A1B6CJV6 E2BXB0 A0A195FX59 A0A1E1WIS7 K7IV24 A0A2J7Q9N9 A0A232FA17 A0A1I8NA52 A0A2A3ENY1 A0A0C9R901 A0A1W4X145 B9W4M6 A0A1I8M708 A0A3L8E0Z4 A0A3G5BIH1 A0A310SIQ6 A0A1Y9J147 Q7PMZ0 A0A182YDC7 W5JIV8 A0A1E1W9K3 A0A182V9C7 A0A023GNA5 A0A1W7RAV1 A0A336L455 A0A224Z6E4 A0A182P9E3 K7IP64 A0A182RFB8 A0A0C9PYP4 J3JW05 A0A1I8N597 A0A131YXB8 A0A026VTG8 A0A0L7QX14 A0A182HNM1 A0A195AUG0 A0A1A9WW87 A0A182JYW3 A0A084WBG2 A0A1I8PQA5 A0A182TR84 W8AS88 A0A088A9Q0 A0A1Y1MRG9 A0A1I8M8P6 A0A154NZ13 A0A293MTR0 B4LGK4 A0A2I9LNU5 A0A067RRG0 A0A0A1WN20 A0A0K8W077 A0A1A9VMJ8 A0A1B0FRI3 A0A1A9Y3F6 T1J5M9 A0A3B0K462 A0A0P4WJ86 A0A1D1UPI1 A0A1B0AEJ3 N6UG27 A0A182MXX8 B4H4A7 Q29DW4 A0A0M5J3G2 A0A1B0ASD3 D6WQS3 B3ML05 A0A034WDZ5
EC Number
3.4.16.-
Pubmed
19121390
22118469
26354079
18362917
19820115
26483478
+ More
20075255 17510324 21347285 20566863 21719571 20798317 28648823 25315136 30249741 30400621 12364791 14747013 17210077 25244985 20920257 23761445 28797301 22516182 26830274 24508170 24438588 24495485 28004739 17994087 29248469 24845553 25830018 27649274 23537049 15632085 25348373
20075255 17510324 21347285 20566863 21719571 20798317 28648823 25315136 30249741 30400621 12364791 14747013 17210077 25244985 20920257 23761445 28797301 22516182 26830274 24508170 24438588 24495485 28004739 17994087 29248469 24845553 25830018 27649274 23537049 15632085 25348373
EMBL
BABH01037195
BABH01037196
GDQN01002481
JAT88573.1
NWSH01000675
PCG74880.1
+ More
NWSH01000162 PCG78925.1 GDQN01002194 GDQN01001899 JAT88860.1 JAT89155.1 AGBW02012825 OWR44390.1 ODYU01006866 SOQ49094.1 KQ459592 KPI96799.1 KQ461194 KPJ06964.1 KQ971312 EEZ98225.1 EEZ98227.1 EEZ98419.1 EEZ98226.2 JXUM01050104 KQ561634 KXJ77929.1 DS231852 EDS37649.1 PCG74881.1 OWR44389.1 PCG78924.1 KPJ06965.1 CH477418 EAT41297.1 ADTU01005771 ADTU01005772 ADTU01005773 DS235815 EEB17508.1 KPI96800.1 GL888378 EGI61995.1 KQ982691 KYQ52053.1 GEDC01023590 JAS13708.1 GL451230 EFN79693.1 KQ981200 KYN45011.1 GDQN01004206 JAT86848.1 AAZX01001182 AAZX01009082 AAZX01018430 NEVH01016346 PNF25303.1 NNAY01000633 OXU27299.1 KZ288212 PBC32839.1 GBYB01012834 JAG82601.1 FM212915 CAR82263.1 QOIP01000001 RLU26384.1 MK075176 AYV99579.1 KQ767283 OAD53440.1 AAAB01008966 EAA13032.5 ADMH02001049 ETN64312.1 GDQN01007435 JAT83619.1 GBBM01000092 JAC35326.1 GFAH01000131 JAV48258.1 UFQS01001511 UFQT01001511 SSX11319.1 SSX30887.1 GFPF01011645 MAA22791.1 GBYB01006648 JAG76415.1 BT127423 AEE62385.1 GEDV01004724 JAP83833.1 KK107965 EZA47037.1 KQ414705 KOC63106.1 KQ976738 KYM75660.1 ATLV01022351 KE525331 KFB47556.1 GAMC01017613 JAB88942.1 GEZM01023580 JAV88292.1 KQ434783 KZC04812.1 GFWV01019464 MAA44192.1 CH940647 EDW69442.1 GFWZ01000068 MBW20058.1 KK852510 KDR22344.1 GBXI01014206 JAD00086.1 GDHF01009476 GDHF01007808 JAI42838.1 JAI44506.1 CCAG010000500 JH431866 OUUW01000012 SPP87492.1 GDRN01085288 JAI61353.1 BDGG01000002 GAU91639.1 APGK01036625 KB740941 KB632409 ENN77592.1 ERL95175.1 CH479208 EDW31222.1 CH379070 EAL30300.2 CP012525 ALC43281.1 JXJN01002732 KQ971354 EFA06031.2 CH902620 EDV30663.1 GAKP01005186 JAC53766.1
NWSH01000162 PCG78925.1 GDQN01002194 GDQN01001899 JAT88860.1 JAT89155.1 AGBW02012825 OWR44390.1 ODYU01006866 SOQ49094.1 KQ459592 KPI96799.1 KQ461194 KPJ06964.1 KQ971312 EEZ98225.1 EEZ98227.1 EEZ98419.1 EEZ98226.2 JXUM01050104 KQ561634 KXJ77929.1 DS231852 EDS37649.1 PCG74881.1 OWR44389.1 PCG78924.1 KPJ06965.1 CH477418 EAT41297.1 ADTU01005771 ADTU01005772 ADTU01005773 DS235815 EEB17508.1 KPI96800.1 GL888378 EGI61995.1 KQ982691 KYQ52053.1 GEDC01023590 JAS13708.1 GL451230 EFN79693.1 KQ981200 KYN45011.1 GDQN01004206 JAT86848.1 AAZX01001182 AAZX01009082 AAZX01018430 NEVH01016346 PNF25303.1 NNAY01000633 OXU27299.1 KZ288212 PBC32839.1 GBYB01012834 JAG82601.1 FM212915 CAR82263.1 QOIP01000001 RLU26384.1 MK075176 AYV99579.1 KQ767283 OAD53440.1 AAAB01008966 EAA13032.5 ADMH02001049 ETN64312.1 GDQN01007435 JAT83619.1 GBBM01000092 JAC35326.1 GFAH01000131 JAV48258.1 UFQS01001511 UFQT01001511 SSX11319.1 SSX30887.1 GFPF01011645 MAA22791.1 GBYB01006648 JAG76415.1 BT127423 AEE62385.1 GEDV01004724 JAP83833.1 KK107965 EZA47037.1 KQ414705 KOC63106.1 KQ976738 KYM75660.1 ATLV01022351 KE525331 KFB47556.1 GAMC01017613 JAB88942.1 GEZM01023580 JAV88292.1 KQ434783 KZC04812.1 GFWV01019464 MAA44192.1 CH940647 EDW69442.1 GFWZ01000068 MBW20058.1 KK852510 KDR22344.1 GBXI01014206 JAD00086.1 GDHF01009476 GDHF01007808 JAI42838.1 JAI44506.1 CCAG010000500 JH431866 OUUW01000012 SPP87492.1 GDRN01085288 JAI61353.1 BDGG01000002 GAU91639.1 APGK01036625 KB740941 KB632409 ENN77592.1 ERL95175.1 CH479208 EDW31222.1 CH379070 EAL30300.2 CP012525 ALC43281.1 JXJN01002732 KQ971354 EFA06031.2 CH902620 EDV30663.1 GAKP01005186 JAC53766.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000007266
+ More
UP000069940 UP000249989 UP000002320 UP000002358 UP000008820 UP000005205 UP000009046 UP000007755 UP000075809 UP000192223 UP000008237 UP000078541 UP000235965 UP000215335 UP000095301 UP000242457 UP000279307 UP000076407 UP000007062 UP000076408 UP000000673 UP000075903 UP000075885 UP000075900 UP000053097 UP000053825 UP000078540 UP000091820 UP000075881 UP000030765 UP000095300 UP000075902 UP000005203 UP000076502 UP000008792 UP000027135 UP000078200 UP000092444 UP000092443 UP000268350 UP000186922 UP000092445 UP000019118 UP000030742 UP000075884 UP000008744 UP000001819 UP000092553 UP000092460 UP000007801
UP000069940 UP000249989 UP000002320 UP000002358 UP000008820 UP000005205 UP000009046 UP000007755 UP000075809 UP000192223 UP000008237 UP000078541 UP000235965 UP000215335 UP000095301 UP000242457 UP000279307 UP000076407 UP000007062 UP000076408 UP000000673 UP000075903 UP000075885 UP000075900 UP000053097 UP000053825 UP000078540 UP000091820 UP000075881 UP000030765 UP000095300 UP000075902 UP000005203 UP000076502 UP000008792 UP000027135 UP000078200 UP000092444 UP000092443 UP000268350 UP000186922 UP000092445 UP000019118 UP000030742 UP000075884 UP000008744 UP000001819 UP000092553 UP000092460 UP000007801
Interpro
IPR001563
Peptidase_S10
+ More
IPR029058 AB_hydrolase
IPR020846 MFS_dom
IPR005829 Sugar_transporter_CS
IPR036259 MFS_trans_sf
IPR005828 MFS_sugar_transport-like
IPR003663 Sugar/inositol_transpt
IPR018202 Ser_caboxypep_ser_AS
IPR016295 Proteasome_beta4
IPR029055 Ntn_hydrolases_N
IPR016050 Proteasome_bsu_CS
IPR001353 Proteasome_sua/b
IPR023333 Proteasome_suB-type
IPR029058 AB_hydrolase
IPR020846 MFS_dom
IPR005829 Sugar_transporter_CS
IPR036259 MFS_trans_sf
IPR005828 MFS_sugar_transport-like
IPR003663 Sugar/inositol_transpt
IPR018202 Ser_caboxypep_ser_AS
IPR016295 Proteasome_beta4
IPR029055 Ntn_hydrolases_N
IPR016050 Proteasome_bsu_CS
IPR001353 Proteasome_sua/b
IPR023333 Proteasome_suB-type
Gene 3D
ProteinModelPortal
H9JLE8
A0A1E1WNG9
A0A2A4JTZ5
A0A2A4K3U2
A0A1E1WQ57
A0A212ESC2
+ More
A0A2H1W9D8 A0A194PVL1 A0A194QNN2 D6W900 D6W8Z8 H9JLE7 D6W901 D6W8Z9 A0A182GA02 B0W744 A0A2A4JS98 A0A212ESD4 A0A2A4K4T6 A0A194QPS0 K7IP63 Q173P0 A0A158P0S0 E0VVV2 A0A194Q057 F4WUY1 A0A151WVZ7 K7J676 A0A1W4XBY5 A0A1B6CJV6 E2BXB0 A0A195FX59 A0A1E1WIS7 K7IV24 A0A2J7Q9N9 A0A232FA17 A0A1I8NA52 A0A2A3ENY1 A0A0C9R901 A0A1W4X145 B9W4M6 A0A1I8M708 A0A3L8E0Z4 A0A3G5BIH1 A0A310SIQ6 A0A1Y9J147 Q7PMZ0 A0A182YDC7 W5JIV8 A0A1E1W9K3 A0A182V9C7 A0A023GNA5 A0A1W7RAV1 A0A336L455 A0A224Z6E4 A0A182P9E3 K7IP64 A0A182RFB8 A0A0C9PYP4 J3JW05 A0A1I8N597 A0A131YXB8 A0A026VTG8 A0A0L7QX14 A0A182HNM1 A0A195AUG0 A0A1A9WW87 A0A182JYW3 A0A084WBG2 A0A1I8PQA5 A0A182TR84 W8AS88 A0A088A9Q0 A0A1Y1MRG9 A0A1I8M8P6 A0A154NZ13 A0A293MTR0 B4LGK4 A0A2I9LNU5 A0A067RRG0 A0A0A1WN20 A0A0K8W077 A0A1A9VMJ8 A0A1B0FRI3 A0A1A9Y3F6 T1J5M9 A0A3B0K462 A0A0P4WJ86 A0A1D1UPI1 A0A1B0AEJ3 N6UG27 A0A182MXX8 B4H4A7 Q29DW4 A0A0M5J3G2 A0A1B0ASD3 D6WQS3 B3ML05 A0A034WDZ5
A0A2H1W9D8 A0A194PVL1 A0A194QNN2 D6W900 D6W8Z8 H9JLE7 D6W901 D6W8Z9 A0A182GA02 B0W744 A0A2A4JS98 A0A212ESD4 A0A2A4K4T6 A0A194QPS0 K7IP63 Q173P0 A0A158P0S0 E0VVV2 A0A194Q057 F4WUY1 A0A151WVZ7 K7J676 A0A1W4XBY5 A0A1B6CJV6 E2BXB0 A0A195FX59 A0A1E1WIS7 K7IV24 A0A2J7Q9N9 A0A232FA17 A0A1I8NA52 A0A2A3ENY1 A0A0C9R901 A0A1W4X145 B9W4M6 A0A1I8M708 A0A3L8E0Z4 A0A3G5BIH1 A0A310SIQ6 A0A1Y9J147 Q7PMZ0 A0A182YDC7 W5JIV8 A0A1E1W9K3 A0A182V9C7 A0A023GNA5 A0A1W7RAV1 A0A336L455 A0A224Z6E4 A0A182P9E3 K7IP64 A0A182RFB8 A0A0C9PYP4 J3JW05 A0A1I8N597 A0A131YXB8 A0A026VTG8 A0A0L7QX14 A0A182HNM1 A0A195AUG0 A0A1A9WW87 A0A182JYW3 A0A084WBG2 A0A1I8PQA5 A0A182TR84 W8AS88 A0A088A9Q0 A0A1Y1MRG9 A0A1I8M8P6 A0A154NZ13 A0A293MTR0 B4LGK4 A0A2I9LNU5 A0A067RRG0 A0A0A1WN20 A0A0K8W077 A0A1A9VMJ8 A0A1B0FRI3 A0A1A9Y3F6 T1J5M9 A0A3B0K462 A0A0P4WJ86 A0A1D1UPI1 A0A1B0AEJ3 N6UG27 A0A182MXX8 B4H4A7 Q29DW4 A0A0M5J3G2 A0A1B0ASD3 D6WQS3 B3ML05 A0A034WDZ5
PDB
1AC5
E-value=2.63523e-15,
Score=201
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
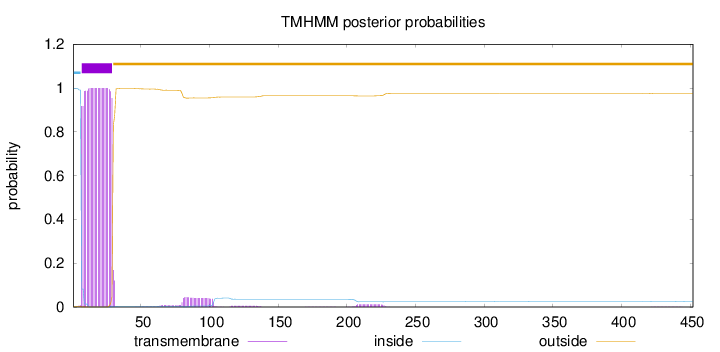
Length:
452
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.5408199999999
Exp number, first 60 AAs:
23.05185
Total prob of N-in:
0.99737
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 452
Population Genetic Test Statistics
Pi
20.505773
Theta
23.764653
Tajima's D
-0.790687
CLR
1.671419
CSRT
0.177941102944853
Interpretation
Uncertain
