Gene
KWMTBOMO07411 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010348
Annotation
PREDICTED:_retinoid-inducible_serine_carboxypeptidase-like_[Amyelois_transitella]
Full name
Carboxypeptidase
Location in the cell
Lysosomal Reliability : 1.6
Sequence
CDS
ATGACGAGTATTCATAAAAAAATGTCGCTTCTCTATTTCTTTTTAGCGGTATTGTTCAGTAAAAGTGTGTCAGGTCAATTCGGTCCGTTTGAACAAGACTTCGGCTATGTGACGGTAAGAGAAGGAGCTCACATGTTTTGGTGGTTATTCTACACAACAGCTCCAGTAGAGAAATATACTGAACGTCCACTCATCGTTTGGTTGCAAGGAGGCCCTGGGGGCTCCTCCACAGGAATTGGAAACTTCGAAATTTTAGGTCCACTCGACTTGTCTTTACAAGAGAGAAATCACAGTTGGGTGAAGAATTTCAATGTTTTGTTCGTTGATAATCCGGTAGGCACTGGCTTTAGTTATGTTGACAATGTGATGCAATTAACTAAGACGAATGATGAAATCGCTCTAGATTTCGTTGAGCTAATGCGTGGATTCTACAATAGACGTCCTGAGTTCAACGATGTTCCTCTCTACATTTACGGTCAGTCGTACGGTGGTAAAATGGCCATTGATATGGGTCTCCGCATGCATGAGGCCGAAAAAGCAGGTACAATACGCTCAAACTTAAAAGGAATAGCGATGGGTAACGCTTGGATATCTCCAGTAGATTCTACTCTCACTTGGGGTCCACTACTGTTAGCTGCTGGCTTAGTAGACCAAGAAGGATATGAGGATATTCAAGCTGCCGCCAGGGAATCCGAGCGATTATTCAATGAAGGACAGTATCTTCAGTCCACGACTCAGTGGGCAAACACACAGCAGGTCGTATTCAGAAGAACTACAAGAGTGGATTTCTACAACATTCTCACTAAAATGGAAGTTTCCGCTTCTGTTCCAGACACTAGATCACCTATTGAACTATCGCGGGATATGTTGATCCGCGATCGACCATACATGTCTCCTGTGAGACAGGATGAATTAAACCTGAACACTTTAATGAACACTCAGGTCAAGGAAGCTCTGAACATTCCAGACCATGTGACATGGGGCTCACAGTCTGGTAATGTTTTTAATATGCTCAGGGAAGATTTCATGAAACCAGTCACAGAAGGAATCGAAAAACTCCTGAATGAAACAGACATTATTTTGACCAAATATAACGGAAATCTAGATTTGATTTGTGATACACCTGGTCAAATTTTATGGGTAGACAGGCTTCGGTGGCCTGGATCGGAAGCTTACAAAAATGCTCCTCGTCTTCCTATTTGGGAAAACAATAAACTAGAAGGTTATTATAAAGCCTTCGAGAATTTTAGATTCTTCTGGATCAATGTGGCAGGACATAGCGTACCAAGGGACAACCCAGAAGGAACCAACGCTTTTCTACGCGATATGACTTCGTTTGGTTAA
Protein
MTSIHKKMSLLYFFLAVLFSKSVSGQFGPFEQDFGYVTVREGAHMFWWLFYTTAPVEKYTERPLIVWLQGGPGGSSTGIGNFEILGPLDLSLQERNHSWVKNFNVLFVDNPVGTGFSYVDNVMQLTKTNDEIALDFVELMRGFYNRRPEFNDVPLYIYGQSYGGKMAIDMGLRMHEAEKAGTIRSNLKGIAMGNAWISPVDSTLTWGPLLLAAGLVDQEGYEDIQAAARESERLFNEGQYLQSTTQWANTQQVVFRRTTRVDFYNILTKMEVSASVPDTRSPIELSRDMLIRDRPYMSPVRQDELNLNTLMNTQVKEALNIPDHVTWGSQSGNVFNMLREDFMKPVTEGIEKLLNETDIILTKYNGNLDLICDTPGQILWVDRLRWPGSEAYKNAPRLPIWENNKLEGYYKAFENFRFFWINVAGHSVPRDNPEGTNAFLRDMTSFG
Summary
Similarity
Belongs to the peptidase S10 family.
Feature
chain Carboxypeptidase
Uniprot
H9JLE7
A0A2A4JS98
A0A1E1WIS7
A0A194QPS0
A0A212ESD4
A0A2A4K4T6
+ More
A0A1E1W9K3 A0A194Q057 A0A0L7LFF7 A0A3L8E0Z4 K7J676 A0A3G5BIH1 A0A034WDZ5 A0A232EW11 A0A195CXH2 A0A0K8W077 A0A0A1WN20 W8AS88 A0A1I8PQA5 B4LGK4 A0A1I8N597 A0A026VTG8 A0A1L8EFY1 A0A1W4VTI2 E2BXB0 A0A1W4W3W2 A0A1J1ISE0 A0A1W4W5A2 A0A336L455 B4PCM1 D6W901 A0A1A9WW87 A0A158P0S0 A0A0Q5UCT9 A0A1A9VMJ8 B3NJA8 A0A0J9RLR9 A0A0L0C047 B4J2S2 A0A1L8EGA9 A0A1B0FRI3 B4QLE2 Q8IRI8 Q5U151 A0A151WVZ7 A0A1A9Y3F6 B3MAE0 A0A1B0AEJ3 B4L0B2 D6W8Z8 A0A1B0ASD3 A0A1L8DPI1 Q29DW4 A0A2A3ENY1 U5EQW7 F4WUY1 B4H4A7 A0A1L8DPK9 B4MLC1 A0A154NZ13 A0A2J7Q9N9 A0A195FX59 A0A0M5J3G2 A0A088A9Q0 A0A182M033 B4MLC0 A0A3B0K462 Q9W0N8 A0A3B0K2E8 A0A1W4VSF9 A0A0J9RLD6 B4J2S3 B4L0B1 B4QLE1 B4PCM0 Q29DW3 A0A182V9C7 B4H4A8 A0A1Y9J147 A0A310SIQ6 Q7PMZ0 B4HVI6 A0A1W4W621 A0A084WBG2 B4LGK5 B3MAD9 B3NJA7 A0A182GA02 A0A1I8M708 B4L0B0 J3JW05 D6W8Z9 A0A1Q3FSI3 A0A1L8EA78 A0A0L7QX14 A0A182MXX8 A0A182HNM1 D6W900 W5JIV8
A0A1E1W9K3 A0A194Q057 A0A0L7LFF7 A0A3L8E0Z4 K7J676 A0A3G5BIH1 A0A034WDZ5 A0A232EW11 A0A195CXH2 A0A0K8W077 A0A0A1WN20 W8AS88 A0A1I8PQA5 B4LGK4 A0A1I8N597 A0A026VTG8 A0A1L8EFY1 A0A1W4VTI2 E2BXB0 A0A1W4W3W2 A0A1J1ISE0 A0A1W4W5A2 A0A336L455 B4PCM1 D6W901 A0A1A9WW87 A0A158P0S0 A0A0Q5UCT9 A0A1A9VMJ8 B3NJA8 A0A0J9RLR9 A0A0L0C047 B4J2S2 A0A1L8EGA9 A0A1B0FRI3 B4QLE2 Q8IRI8 Q5U151 A0A151WVZ7 A0A1A9Y3F6 B3MAE0 A0A1B0AEJ3 B4L0B2 D6W8Z8 A0A1B0ASD3 A0A1L8DPI1 Q29DW4 A0A2A3ENY1 U5EQW7 F4WUY1 B4H4A7 A0A1L8DPK9 B4MLC1 A0A154NZ13 A0A2J7Q9N9 A0A195FX59 A0A0M5J3G2 A0A088A9Q0 A0A182M033 B4MLC0 A0A3B0K462 Q9W0N8 A0A3B0K2E8 A0A1W4VSF9 A0A0J9RLD6 B4J2S3 B4L0B1 B4QLE1 B4PCM0 Q29DW3 A0A182V9C7 B4H4A8 A0A1Y9J147 A0A310SIQ6 Q7PMZ0 B4HVI6 A0A1W4W621 A0A084WBG2 B4LGK5 B3MAD9 B3NJA7 A0A182GA02 A0A1I8M708 B4L0B0 J3JW05 D6W8Z9 A0A1Q3FSI3 A0A1L8EA78 A0A0L7QX14 A0A182MXX8 A0A182HNM1 D6W900 W5JIV8
EC Number
3.4.16.-
Pubmed
19121390
26354079
22118469
26227816
30249741
20075255
+ More
30400621 25348373 28648823 25830018 24495485 17994087 25315136 24508170 20798317 17550304 18362917 19820115 21347285 22936249 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 21719571 26109357 26109356 12364791 14747013 17210077 24438588 26483478 22516182 20920257 23761445
30400621 25348373 28648823 25830018 24495485 17994087 25315136 24508170 20798317 17550304 18362917 19820115 21347285 22936249 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 21719571 26109357 26109356 12364791 14747013 17210077 24438588 26483478 22516182 20920257 23761445
EMBL
BABH01037195
NWSH01000675
PCG74881.1
GDQN01004206
JAT86848.1
KQ461194
+ More
KPJ06965.1 AGBW02012825 OWR44389.1 NWSH01000162 PCG78924.1 GDQN01007435 JAT83619.1 KQ459592 KPI96800.1 JTDY01001329 KOB74182.1 QOIP01000001 RLU26384.1 MK075176 AYV99579.1 GAKP01005186 JAC53766.1 NNAY01001926 OXU22542.1 KQ977141 KYN05363.1 GDHF01009476 GDHF01007808 JAI42838.1 JAI44506.1 GBXI01014206 JAD00086.1 GAMC01017613 JAB88942.1 CH940647 EDW69442.1 KK107965 EZA47037.1 GFDG01001169 JAV17630.1 GL451230 EFN79693.1 CVRI01000059 CRL03157.1 UFQS01001511 UFQT01001511 SSX11319.1 SSX30887.1 CM000159 EDW92744.1 KQ971312 EEZ98419.1 ADTU01005771 ADTU01005772 ADTU01005773 CH954178 KQS43041.1 EDV50070.1 CM002912 KMY96707.1 JRES01001084 KNC25675.1 CH916366 EDV96063.1 GFDG01001160 JAV17639.1 CCAG010000500 CM000363 EDX08715.1 AE014296 AAN11449.1 BT016041 AAV36926.1 KQ982691 KYQ52053.1 CH902618 EDV40191.2 CH933809 EDW18058.2 EEZ98227.1 JXJN01002732 GFDF01005800 JAV08284.1 CH379070 EAL30300.2 KZ288212 PBC32839.1 GANO01003086 JAB56785.1 GL888378 EGI61995.1 CH479208 EDW31222.1 GFDF01005799 JAV08285.1 CH963847 EDW73379.1 KQ434783 KZC04812.1 NEVH01016346 PNF25303.1 KQ981200 KYN45011.1 CP012525 ALC43281.1 AXCM01001232 EDW73378.1 OUUW01000012 SPP87492.1 AY051562 AAF47405.1 AAK92986.1 SPP87493.1 KMY96706.1 EDV96064.1 EDW18057.1 EDX08714.1 EDW92743.1 EAL30301.1 EDW31223.1 KQ767283 OAD53440.1 AAAB01008966 EAA13032.5 CH480817 EDW49951.1 ATLV01022351 KE525331 KFB47556.1 EDW69443.1 EDV40190.1 EDV50069.1 JXUM01050104 KQ561634 KXJ77929.1 EDW18056.1 BT127423 AEE62385.1 EEZ98226.2 GFDL01004491 JAV30554.1 GFDG01003280 JAV15519.1 KQ414705 KOC63106.1 EEZ98225.1 ADMH02001049 ETN64312.1
KPJ06965.1 AGBW02012825 OWR44389.1 NWSH01000162 PCG78924.1 GDQN01007435 JAT83619.1 KQ459592 KPI96800.1 JTDY01001329 KOB74182.1 QOIP01000001 RLU26384.1 MK075176 AYV99579.1 GAKP01005186 JAC53766.1 NNAY01001926 OXU22542.1 KQ977141 KYN05363.1 GDHF01009476 GDHF01007808 JAI42838.1 JAI44506.1 GBXI01014206 JAD00086.1 GAMC01017613 JAB88942.1 CH940647 EDW69442.1 KK107965 EZA47037.1 GFDG01001169 JAV17630.1 GL451230 EFN79693.1 CVRI01000059 CRL03157.1 UFQS01001511 UFQT01001511 SSX11319.1 SSX30887.1 CM000159 EDW92744.1 KQ971312 EEZ98419.1 ADTU01005771 ADTU01005772 ADTU01005773 CH954178 KQS43041.1 EDV50070.1 CM002912 KMY96707.1 JRES01001084 KNC25675.1 CH916366 EDV96063.1 GFDG01001160 JAV17639.1 CCAG010000500 CM000363 EDX08715.1 AE014296 AAN11449.1 BT016041 AAV36926.1 KQ982691 KYQ52053.1 CH902618 EDV40191.2 CH933809 EDW18058.2 EEZ98227.1 JXJN01002732 GFDF01005800 JAV08284.1 CH379070 EAL30300.2 KZ288212 PBC32839.1 GANO01003086 JAB56785.1 GL888378 EGI61995.1 CH479208 EDW31222.1 GFDF01005799 JAV08285.1 CH963847 EDW73379.1 KQ434783 KZC04812.1 NEVH01016346 PNF25303.1 KQ981200 KYN45011.1 CP012525 ALC43281.1 AXCM01001232 EDW73378.1 OUUW01000012 SPP87492.1 AY051562 AAF47405.1 AAK92986.1 SPP87493.1 KMY96706.1 EDV96064.1 EDW18057.1 EDX08714.1 EDW92743.1 EAL30301.1 EDW31223.1 KQ767283 OAD53440.1 AAAB01008966 EAA13032.5 CH480817 EDW49951.1 ATLV01022351 KE525331 KFB47556.1 EDW69443.1 EDV40190.1 EDV50069.1 JXUM01050104 KQ561634 KXJ77929.1 EDW18056.1 BT127423 AEE62385.1 EEZ98226.2 GFDL01004491 JAV30554.1 GFDG01003280 JAV15519.1 KQ414705 KOC63106.1 EEZ98225.1 ADMH02001049 ETN64312.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000037510
+ More
UP000279307 UP000002358 UP000215335 UP000078542 UP000095300 UP000008792 UP000095301 UP000053097 UP000192221 UP000008237 UP000183832 UP000002282 UP000007266 UP000091820 UP000005205 UP000008711 UP000078200 UP000037069 UP000001070 UP000092444 UP000000304 UP000000803 UP000075809 UP000092443 UP000007801 UP000092445 UP000009192 UP000092460 UP000001819 UP000242457 UP000007755 UP000008744 UP000007798 UP000076502 UP000235965 UP000078541 UP000092553 UP000005203 UP000075883 UP000268350 UP000075903 UP000076407 UP000007062 UP000001292 UP000030765 UP000069940 UP000249989 UP000053825 UP000075884 UP000000673
UP000279307 UP000002358 UP000215335 UP000078542 UP000095300 UP000008792 UP000095301 UP000053097 UP000192221 UP000008237 UP000183832 UP000002282 UP000007266 UP000091820 UP000005205 UP000008711 UP000078200 UP000037069 UP000001070 UP000092444 UP000000304 UP000000803 UP000075809 UP000092443 UP000007801 UP000092445 UP000009192 UP000092460 UP000001819 UP000242457 UP000007755 UP000008744 UP000007798 UP000076502 UP000235965 UP000078541 UP000092553 UP000005203 UP000075883 UP000268350 UP000075903 UP000076407 UP000007062 UP000001292 UP000030765 UP000069940 UP000249989 UP000053825 UP000075884 UP000000673
Pfam
PF00450 Peptidase_S10
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9JLE7
A0A2A4JS98
A0A1E1WIS7
A0A194QPS0
A0A212ESD4
A0A2A4K4T6
+ More
A0A1E1W9K3 A0A194Q057 A0A0L7LFF7 A0A3L8E0Z4 K7J676 A0A3G5BIH1 A0A034WDZ5 A0A232EW11 A0A195CXH2 A0A0K8W077 A0A0A1WN20 W8AS88 A0A1I8PQA5 B4LGK4 A0A1I8N597 A0A026VTG8 A0A1L8EFY1 A0A1W4VTI2 E2BXB0 A0A1W4W3W2 A0A1J1ISE0 A0A1W4W5A2 A0A336L455 B4PCM1 D6W901 A0A1A9WW87 A0A158P0S0 A0A0Q5UCT9 A0A1A9VMJ8 B3NJA8 A0A0J9RLR9 A0A0L0C047 B4J2S2 A0A1L8EGA9 A0A1B0FRI3 B4QLE2 Q8IRI8 Q5U151 A0A151WVZ7 A0A1A9Y3F6 B3MAE0 A0A1B0AEJ3 B4L0B2 D6W8Z8 A0A1B0ASD3 A0A1L8DPI1 Q29DW4 A0A2A3ENY1 U5EQW7 F4WUY1 B4H4A7 A0A1L8DPK9 B4MLC1 A0A154NZ13 A0A2J7Q9N9 A0A195FX59 A0A0M5J3G2 A0A088A9Q0 A0A182M033 B4MLC0 A0A3B0K462 Q9W0N8 A0A3B0K2E8 A0A1W4VSF9 A0A0J9RLD6 B4J2S3 B4L0B1 B4QLE1 B4PCM0 Q29DW3 A0A182V9C7 B4H4A8 A0A1Y9J147 A0A310SIQ6 Q7PMZ0 B4HVI6 A0A1W4W621 A0A084WBG2 B4LGK5 B3MAD9 B3NJA7 A0A182GA02 A0A1I8M708 B4L0B0 J3JW05 D6W8Z9 A0A1Q3FSI3 A0A1L8EA78 A0A0L7QX14 A0A182MXX8 A0A182HNM1 D6W900 W5JIV8
A0A1E1W9K3 A0A194Q057 A0A0L7LFF7 A0A3L8E0Z4 K7J676 A0A3G5BIH1 A0A034WDZ5 A0A232EW11 A0A195CXH2 A0A0K8W077 A0A0A1WN20 W8AS88 A0A1I8PQA5 B4LGK4 A0A1I8N597 A0A026VTG8 A0A1L8EFY1 A0A1W4VTI2 E2BXB0 A0A1W4W3W2 A0A1J1ISE0 A0A1W4W5A2 A0A336L455 B4PCM1 D6W901 A0A1A9WW87 A0A158P0S0 A0A0Q5UCT9 A0A1A9VMJ8 B3NJA8 A0A0J9RLR9 A0A0L0C047 B4J2S2 A0A1L8EGA9 A0A1B0FRI3 B4QLE2 Q8IRI8 Q5U151 A0A151WVZ7 A0A1A9Y3F6 B3MAE0 A0A1B0AEJ3 B4L0B2 D6W8Z8 A0A1B0ASD3 A0A1L8DPI1 Q29DW4 A0A2A3ENY1 U5EQW7 F4WUY1 B4H4A7 A0A1L8DPK9 B4MLC1 A0A154NZ13 A0A2J7Q9N9 A0A195FX59 A0A0M5J3G2 A0A088A9Q0 A0A182M033 B4MLC0 A0A3B0K462 Q9W0N8 A0A3B0K2E8 A0A1W4VSF9 A0A0J9RLD6 B4J2S3 B4L0B1 B4QLE1 B4PCM0 Q29DW3 A0A182V9C7 B4H4A8 A0A1Y9J147 A0A310SIQ6 Q7PMZ0 B4HVI6 A0A1W4W621 A0A084WBG2 B4LGK5 B3MAD9 B3NJA7 A0A182GA02 A0A1I8M708 B4L0B0 J3JW05 D6W8Z9 A0A1Q3FSI3 A0A1L8EA78 A0A0L7QX14 A0A182MXX8 A0A182HNM1 D6W900 W5JIV8
PDB
1IVY
E-value=2.27409e-18,
Score=227
Ontologies
GO
PANTHER
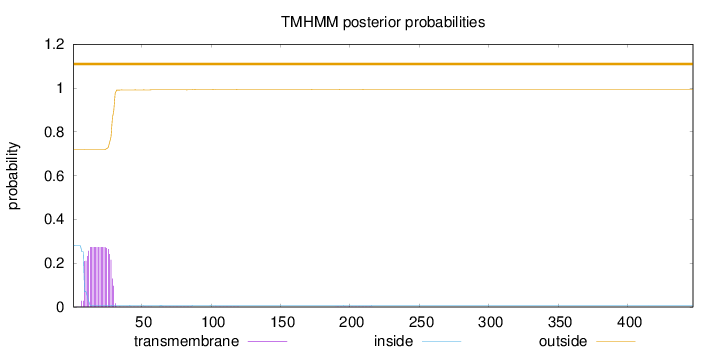
Topology
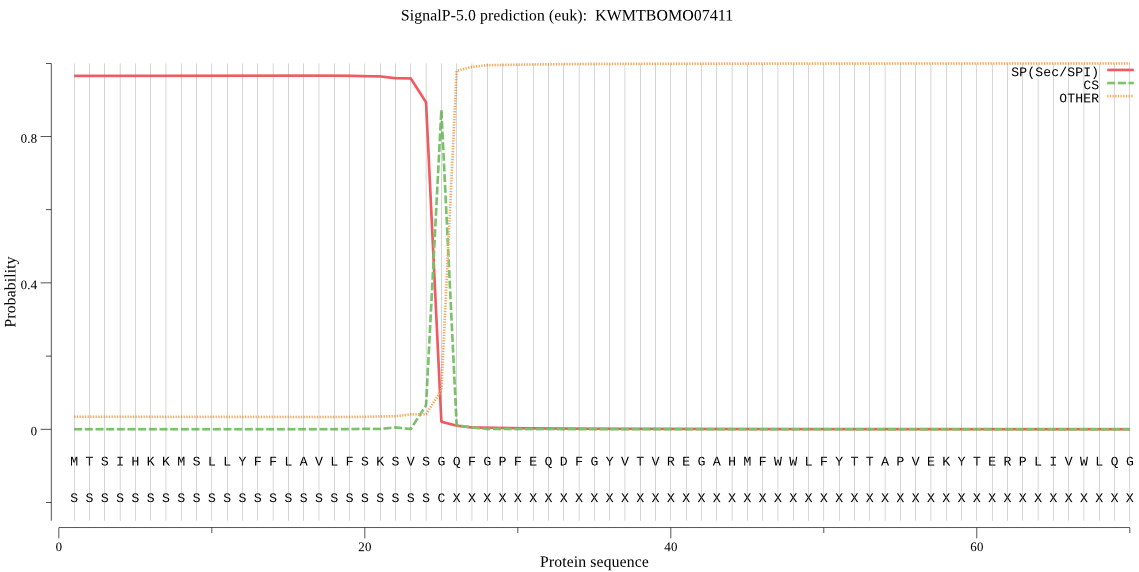
SignalP
Position: 1 - 25,
Likelihood: 0.966050
Length:
447
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.55147
Exp number, first 60 AAs:
5.50766
Total prob of N-in:
0.28015
outside
1 - 447
Population Genetic Test Statistics
Pi
32.3689
Theta
23.949576
Tajima's D
1.128547
CLR
0.602084
CSRT
0.699615019249038
Interpretation
Uncertain
