Gene
KWMTBOMO07409
Pre Gene Modal
BGIBMGA010320
Annotation
PREDICTED:_cuticular_protein_RR-2_motif_140_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.849
Sequence
CDS
ATGATTTTATCAACTTTCTTTATTAGTATATTGGCTTGGTCAGCAATCCGTTGTGAACCACCAGTAAACAGTTACCTACCTCCTTCAAATAGTGCAAACGGAAGGCCTTCATCTCAGTATGGTCCTCCCGGTTCAGGAGGTGCTCCTGGACATAATCAACAAGGTTCAGGAGCTCAATTTAGTCCCAGTAACCAACCCCAGAGCTCTTATCTGCCTCCTAGTCGTTCTGGTCAATCCAGTGTAGACACACAATATGGCCCACCTTCTTCCGGAGGCTTTGGTTCGGGGTCATCTTCTCAAGGCCAACAACTACGCCAGGGTTCCGGACAAGCAAAGCCAGACTCACAATATGGAACTCCAAATCAAAATGGAAGATTCTCTTCGGGATTGACCGGTGGACATTCCCAACCTGGTGCCAGTAGTAATAGTCATCCGACAAGCAATTCTCAGGGCAGACAATCTTTTGGAAATGGCGGATCTTCTAATGGCGGTAGCTTTGGACCAGATAATCAGAGTTTTGATCAGAAGCCTGGTAGTTCTTATAATCCTTCTACTGCTGGATTTCGGTCTTCCAATTCGGATTCAAATTCTAATGGAAACTTCGGTAGTCATGCCGGTGGAGCTCGTTTGACCGATGGTTCTCAGAGTAGTTTTAGTGGAACACCATCCTCTTCGTATGGAACGCCTGGATTCGGAGGTAACGGTGCTAGTCATGGTAATGGCGGCGGCAGTGGTTCAGGTTTTGGAGGAGGAAATGGAGGTTTTGGAGGAGATGACAATAATGAACCAGCAAAATACCAGTTTAGTTATGACGTGGACGATGAACAGACAGGTACTAAATTTGGTCATTCAGAACAGCGAGATGGCGATCTCGCCACTGGGGAGTATAACGTAGTGTTGCCAGATGGCAGGAAGCAGGTTGTTGAGTACGAGGCTGGCCTGGAAGGATACAAGCCTCAGATTCGATATGAGGGTACTGGAAATGGGGGGTCAAGGCAAGGATCTGGATCGGGTGGTTTCGGGGAATCGCAAAGATTGAGTCAAGGATACACTGAAAATGGTCCCAATGAAGGAGGTTTTCAGAGCAACAACGTTGATAGTTTTGGAGGATCAGGAGAACAGACATTTGGTCAATTCCGAGGCTCTAATGCTCAAGCCGGTAATGGATATCCCAGTGGTGGACAGAATGGCCAAAATCAAGGATACCCAAATGGAGGTTCCGGACAAAACGGGAATCAAGGTTTTTCATCGGGTAATAACGGCGCAAATAGACTTAATGGAGGAAGCCAGGGCTATCCAAGTGGCGGACCACGAGGAAATGGTCGAGGGTCTGCCGCTGACCCAGGTAACTTTTCTAATGCCAGCGGAAGAGGAAGCTCTCGAAACTCAGGTGGTACTTCTGGAAGTGATGGTTACCCAAGTGGTGGACCTAATGGGCTTAGAGGCTCTGGATATTAA
Protein
MILSTFFISILAWSAIRCEPPVNSYLPPSNSANGRPSSQYGPPGSGGAPGHNQQGSGAQFSPSNQPQSSYLPPSRSGQSSVDTQYGPPSSGGFGSGSSSQGQQLRQGSGQAKPDSQYGTPNQNGRFSSGLTGGHSQPGASSNSHPTSNSQGRQSFGNGGSSNGGSFGPDNQSFDQKPGSSYNPSTAGFRSSNSDSNSNGNFGSHAGGARLTDGSQSSFSGTPSSSYGTPGFGGNGASHGNGGGSGSGFGGGNGGFGGDDNNEPAKYQFSYDVDDEQTGTKFGHSEQRDGDLATGEYNVVLPDGRKQVVEYEAGLEGYKPQIRYEGTGNGGSRQGSGSGGFGESQRLSQGYTENGPNEGGFQSNNVDSFGGSGEQTFGQFRGSNAQAGNGYPSGGQNGQNQGYPNGGSGQNGNQGFSSGNNGANRLNGGSQGYPSGGPRGNGRGSAADPGNFSNASGRGSSRNSGGTSGSDGYPSGGPNGLRGSGY
Summary
Uniprot
Proteomes
PRIDE
Pfam
PF00379 Chitin_bind_4
ProteinModelPortal
Ontologies
GO
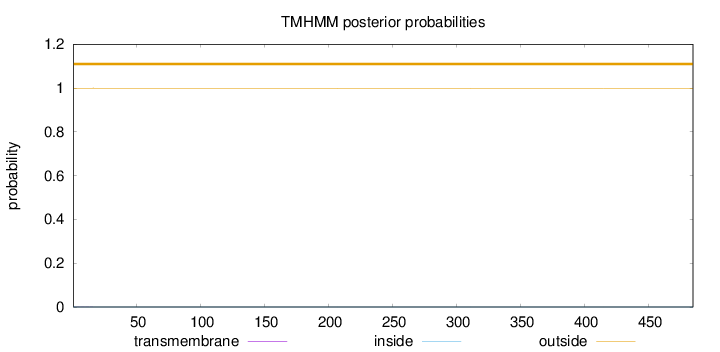
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.976277

Length:
485
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00485
Exp number, first 60 AAs:
0.00485
Total prob of N-in:
0.00074
outside
1 - 485
Population Genetic Test Statistics
Pi
29.730352
Theta
22.78352
Tajima's D
0.871593
CLR
0.483263
CSRT
0.632268386580671
Interpretation
Uncertain
