Gene
KWMTBOMO07407
Pre Gene Modal
BGIBMGA010346
Annotation
PREDICTED:_uncharacterized_protein_LOC106132307_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 2.348
Sequence
CDS
ATGTTAAGTCTTTTGTCACGATCCGAATTGTCTCCAGATTACGGCGTGACAAGCCTCGAACTATTAGAGCTACGGGTCCCGGCGCATGTCCCACTTGGCACCCGAGCTGAACTTTCCTGTAGATGGCAACTCGGCCCGGCCGACATCCTTTATTCCGTCAAATGGTATAAAGACGGAAAGGAATTCTTCAGACACGTGCCACGAGATCAAGAGCCACGAAGAAAATTCCCTCTCCCGGGAGTCGATGTTGAAAAGTCGAGCACGAATGGCGCGAACGTGACTCTCGCCCCGGCCATCCTGGAAACGGGCGGGCGCTACCGCTGCGAGGTGTCCGGGGAACGGCCGCTCTTCCCGACAGTGTCCGACCACGCTGATATGATCATCGTAGCGTTACCAGACCAGGGACCTACGATAACTGGATCAAAAATCCGGTATCAAATCGGCGACCGGGTGCAGGTGAACTGCACGTCAGGCCGGTCCAGGCCCGCCACCAGGCTGGCTTGGTACATCAACGGAGAGCCGGCGCCTACCGCCGCCTTACTGACTCCAACCCACCAAGTTCATAGCGACGGATTGGAGACGACCAGTTTGGGGCTCGATTTCAAAGTGAAATCAAAGCATTTTAGGAAAGGAGACTTAAAATTAAAATGTCTAGCAACAATAGCAACGGTATACTGGAGAAGCAATGAAGAGAGTGTTCAGGGAGAGAAACTGAAAGGTTATTCTAGGTCTCGGGAGCTGACCGAGGGTGCTTCCCGAGCTGATCGCGTCCAAGCAGCAGGAAAATTAGGGGGCTCTGCCCCCGCGCTTCAGCTAACCCACTGTTTACTAATATTCAGTGCTACATTACTACAAACGCTACCAACACGATTACACGTGGACTTTGTTTAA
Protein
MLSLLSRSELSPDYGVTSLELLELRVPAHVPLGTRAELSCRWQLGPADILYSVKWYKDGKEFFRHVPRDQEPRRKFPLPGVDVEKSSTNGANVTLAPAILETGGRYRCEVSGERPLFPTVSDHADMIIVALPDQGPTITGSKIRYQIGDRVQVNCTSGRSRPATRLAWYINGEPAPTAALLTPTHQVHSDGLETTSLGLDFKVKSKHFRKGDLKLKCLATIATVYWRSNEESVQGEKLKGYSRSRELTEGASRADRVQAAGKLGGSAPALQLTHCLLIFSATLLQTLPTRLHVDFV
Summary
Uniprot
H9JLE5
A0A2A4K4S6
A0A2H1V2Q2
A0A212EL69
A0A194R800
A0A194PTQ5
+ More
A0A194PTZ9 A0A182IRE1 A0A1B6D8N3 A0A182RBL5 A0A182NE34 A0A3F2YYX2 A0A182HV74 A0A1B6DEL1 A0A067R8E3 A0A084VP25 A0A182L227 A0A182WT79 A0A182PJ74 A0A182NE33 A0A182JJD3 A0A1B6GB06 W5JNY9 A0A182SIT5 A0A182F4L2 A0A3S2NW58 W5JJP2 B4I589 A0A182K507 A0A084VP22 A0A0J9TPX9 Q9VJF1 A0A1B6CQX7 A0A182RBL4 Q7Q3L1 A0A182W9N1 A0A182M0V6 A0A182WT78 A0A1S4GWR5 A0A182HV73 A0A182G7K8 A0A182L217 A0A182Q4C0 A0A182THU0 A0A182K665 A0A182V637 A0A1Y1JZU7 A0A182YGU0 A0A1Y1K2U9 B3NLE4 A0A182F4L1 A0A182YGU1 A0A1Y1JZV1 A0A224XHL9 B4P8T4 A0A182PJ76 A0A182H6X9 A0A0J9R3R3 A0A182MNW7 Q7Q3L0 Q8SY13 A0A0Q5VUM4 A0A0R1DNZ4 A0A0A1XKB7 A0A2P8Z7Y1 W8B0U9 B4MTM9 B3MMJ5 A0A182PJ75 A0A1S4F2H1 A0A0K8UDR0 A0A3B0KAP3 Q17HG6 A0A182QG80 A0A1A9VL65 A0A3B0K2K1 A0A0P9BMX5 A0A034V4K5 A0A182W9N2 A0A1A9WUN8 B0X9B9 W8BCG7 A0A1I8P4K3 A0A0R3NUA0 Q29L84 A0A0K8VBJ8 A0A1B0A449 A0A182GT17 A0A182WT77 A0A182HV72 Q17HG5 B4JD11 A0A139WK34 A0A1I8P4R7 A0A1S4F2K6 A0A1S4GWQ7 K7J6Q0 A0A1B0AWA0 B4MDF2 A0A1A9YBP9
A0A194PTZ9 A0A182IRE1 A0A1B6D8N3 A0A182RBL5 A0A182NE34 A0A3F2YYX2 A0A182HV74 A0A1B6DEL1 A0A067R8E3 A0A084VP25 A0A182L227 A0A182WT79 A0A182PJ74 A0A182NE33 A0A182JJD3 A0A1B6GB06 W5JNY9 A0A182SIT5 A0A182F4L2 A0A3S2NW58 W5JJP2 B4I589 A0A182K507 A0A084VP22 A0A0J9TPX9 Q9VJF1 A0A1B6CQX7 A0A182RBL4 Q7Q3L1 A0A182W9N1 A0A182M0V6 A0A182WT78 A0A1S4GWR5 A0A182HV73 A0A182G7K8 A0A182L217 A0A182Q4C0 A0A182THU0 A0A182K665 A0A182V637 A0A1Y1JZU7 A0A182YGU0 A0A1Y1K2U9 B3NLE4 A0A182F4L1 A0A182YGU1 A0A1Y1JZV1 A0A224XHL9 B4P8T4 A0A182PJ76 A0A182H6X9 A0A0J9R3R3 A0A182MNW7 Q7Q3L0 Q8SY13 A0A0Q5VUM4 A0A0R1DNZ4 A0A0A1XKB7 A0A2P8Z7Y1 W8B0U9 B4MTM9 B3MMJ5 A0A182PJ75 A0A1S4F2H1 A0A0K8UDR0 A0A3B0KAP3 Q17HG6 A0A182QG80 A0A1A9VL65 A0A3B0K2K1 A0A0P9BMX5 A0A034V4K5 A0A182W9N2 A0A1A9WUN8 B0X9B9 W8BCG7 A0A1I8P4K3 A0A0R3NUA0 Q29L84 A0A0K8VBJ8 A0A1B0A449 A0A182GT17 A0A182WT77 A0A182HV72 Q17HG5 B4JD11 A0A139WK34 A0A1I8P4R7 A0A1S4F2K6 A0A1S4GWQ7 K7J6Q0 A0A1B0AWA0 B4MDF2 A0A1A9YBP9
Pubmed
19121390
22118469
26354079
12364791
24845553
24438588
+ More
20966253 20920257 23761445 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26483478 28004739 25244985 17550304 26109357 26109356 25830018 29403074 24495485 17510324 25348373 15632085 23185243 18362917 19820115 20075255
20966253 20920257 23761445 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26483478 28004739 25244985 17550304 26109357 26109356 25830018 29403074 24495485 17510324 25348373 15632085 23185243 18362917 19820115 20075255
EMBL
BABH01037186
BABH01037187
NWSH01000162
PCG78914.1
ODYU01000403
SOQ35125.1
+ More
AGBW02014125 OWR42236.1 KQ460845 KPJ11996.1 KQ459592 KPI96796.1 KPI96792.1 AXCP01007901 GEDC01015245 JAS22053.1 AAAB01008964 APCN01002496 GEDC01018948 GEDC01013263 JAS18350.1 JAS24035.1 KK852813 KDR15839.1 ATLV01014967 KE524999 KFB39719.1 GECZ01010142 JAS59627.1 ADMH02001006 ETN64469.1 RSAL01000139 RVE46139.1 ADMH02001293 ETN63125.1 CH480822 EDW55545.1 ATLV01014966 KFB39716.1 CM002910 KMY90660.1 AE014134 AAF53598.2 GEDC01021342 JAS15956.1 EAA12864.3 AXCM01002897 APCN01002495 JXUM01046553 KQ561481 KXJ78444.1 AXCN02000872 GEZM01096543 JAV54819.1 GEZM01096542 JAV54821.1 CH954179 EDV54860.1 GEZM01096541 JAV54822.1 GFTR01004439 JAW11987.1 CM000158 EDW90192.1 JXUM01115440 KQ565880 KXJ70572.1 KMY90661.1 AXCM01003227 EAA12775.4 AY075458 AAF53600.2 AAL68271.1 KQS61977.1 KRJ99018.1 GBXI01002905 JAD11387.1 PYGN01000156 PSN52590.1 GAMC01011855 GAMC01011853 JAB94700.1 CH963852 EDW75468.2 CH902620 EDV30941.1 GDHF01027611 JAI24703.1 OUUW01000006 SPP82111.1 CH477249 EAT46096.1 SPP82110.1 KPU73150.1 GAKP01021493 JAC37459.1 DS232530 EDS43051.1 GAMC01011857 JAB94698.1 CH379061 KRT04570.1 EAL32940.2 KRT04569.1 KRT04571.1 GDHF01016071 JAI36243.1 JXUM01017467 KQ560484 KXJ82184.1 EAT46097.1 CH916368 EDW04255.1 KQ971338 KYB28177.1 JXJN01004581 CH940661 EDW71213.2
AGBW02014125 OWR42236.1 KQ460845 KPJ11996.1 KQ459592 KPI96796.1 KPI96792.1 AXCP01007901 GEDC01015245 JAS22053.1 AAAB01008964 APCN01002496 GEDC01018948 GEDC01013263 JAS18350.1 JAS24035.1 KK852813 KDR15839.1 ATLV01014967 KE524999 KFB39719.1 GECZ01010142 JAS59627.1 ADMH02001006 ETN64469.1 RSAL01000139 RVE46139.1 ADMH02001293 ETN63125.1 CH480822 EDW55545.1 ATLV01014966 KFB39716.1 CM002910 KMY90660.1 AE014134 AAF53598.2 GEDC01021342 JAS15956.1 EAA12864.3 AXCM01002897 APCN01002495 JXUM01046553 KQ561481 KXJ78444.1 AXCN02000872 GEZM01096543 JAV54819.1 GEZM01096542 JAV54821.1 CH954179 EDV54860.1 GEZM01096541 JAV54822.1 GFTR01004439 JAW11987.1 CM000158 EDW90192.1 JXUM01115440 KQ565880 KXJ70572.1 KMY90661.1 AXCM01003227 EAA12775.4 AY075458 AAF53600.2 AAL68271.1 KQS61977.1 KRJ99018.1 GBXI01002905 JAD11387.1 PYGN01000156 PSN52590.1 GAMC01011855 GAMC01011853 JAB94700.1 CH963852 EDW75468.2 CH902620 EDV30941.1 GDHF01027611 JAI24703.1 OUUW01000006 SPP82111.1 CH477249 EAT46096.1 SPP82110.1 KPU73150.1 GAKP01021493 JAC37459.1 DS232530 EDS43051.1 GAMC01011857 JAB94698.1 CH379061 KRT04570.1 EAL32940.2 KRT04569.1 KRT04571.1 GDHF01016071 JAI36243.1 JXUM01017467 KQ560484 KXJ82184.1 EAT46097.1 CH916368 EDW04255.1 KQ971338 KYB28177.1 JXJN01004581 CH940661 EDW71213.2
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000075880
+ More
UP000075900 UP000075884 UP000075840 UP000027135 UP000030765 UP000075882 UP000076407 UP000075885 UP000000673 UP000075901 UP000069272 UP000283053 UP000001292 UP000075881 UP000000803 UP000007062 UP000075920 UP000075883 UP000069940 UP000249989 UP000075886 UP000075902 UP000075903 UP000076408 UP000008711 UP000002282 UP000245037 UP000007798 UP000007801 UP000268350 UP000008820 UP000078200 UP000091820 UP000002320 UP000095300 UP000001819 UP000092445 UP000001070 UP000007266 UP000002358 UP000092460 UP000008792 UP000092443
UP000075900 UP000075884 UP000075840 UP000027135 UP000030765 UP000075882 UP000076407 UP000075885 UP000000673 UP000075901 UP000069272 UP000283053 UP000001292 UP000075881 UP000000803 UP000007062 UP000075920 UP000075883 UP000069940 UP000249989 UP000075886 UP000075902 UP000075903 UP000076408 UP000008711 UP000002282 UP000245037 UP000007798 UP000007801 UP000268350 UP000008820 UP000078200 UP000091820 UP000002320 UP000095300 UP000001819 UP000092445 UP000001070 UP000007266 UP000002358 UP000092460 UP000008792 UP000092443
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
H9JLE5
A0A2A4K4S6
A0A2H1V2Q2
A0A212EL69
A0A194R800
A0A194PTQ5
+ More
A0A194PTZ9 A0A182IRE1 A0A1B6D8N3 A0A182RBL5 A0A182NE34 A0A3F2YYX2 A0A182HV74 A0A1B6DEL1 A0A067R8E3 A0A084VP25 A0A182L227 A0A182WT79 A0A182PJ74 A0A182NE33 A0A182JJD3 A0A1B6GB06 W5JNY9 A0A182SIT5 A0A182F4L2 A0A3S2NW58 W5JJP2 B4I589 A0A182K507 A0A084VP22 A0A0J9TPX9 Q9VJF1 A0A1B6CQX7 A0A182RBL4 Q7Q3L1 A0A182W9N1 A0A182M0V6 A0A182WT78 A0A1S4GWR5 A0A182HV73 A0A182G7K8 A0A182L217 A0A182Q4C0 A0A182THU0 A0A182K665 A0A182V637 A0A1Y1JZU7 A0A182YGU0 A0A1Y1K2U9 B3NLE4 A0A182F4L1 A0A182YGU1 A0A1Y1JZV1 A0A224XHL9 B4P8T4 A0A182PJ76 A0A182H6X9 A0A0J9R3R3 A0A182MNW7 Q7Q3L0 Q8SY13 A0A0Q5VUM4 A0A0R1DNZ4 A0A0A1XKB7 A0A2P8Z7Y1 W8B0U9 B4MTM9 B3MMJ5 A0A182PJ75 A0A1S4F2H1 A0A0K8UDR0 A0A3B0KAP3 Q17HG6 A0A182QG80 A0A1A9VL65 A0A3B0K2K1 A0A0P9BMX5 A0A034V4K5 A0A182W9N2 A0A1A9WUN8 B0X9B9 W8BCG7 A0A1I8P4K3 A0A0R3NUA0 Q29L84 A0A0K8VBJ8 A0A1B0A449 A0A182GT17 A0A182WT77 A0A182HV72 Q17HG5 B4JD11 A0A139WK34 A0A1I8P4R7 A0A1S4F2K6 A0A1S4GWQ7 K7J6Q0 A0A1B0AWA0 B4MDF2 A0A1A9YBP9
A0A194PTZ9 A0A182IRE1 A0A1B6D8N3 A0A182RBL5 A0A182NE34 A0A3F2YYX2 A0A182HV74 A0A1B6DEL1 A0A067R8E3 A0A084VP25 A0A182L227 A0A182WT79 A0A182PJ74 A0A182NE33 A0A182JJD3 A0A1B6GB06 W5JNY9 A0A182SIT5 A0A182F4L2 A0A3S2NW58 W5JJP2 B4I589 A0A182K507 A0A084VP22 A0A0J9TPX9 Q9VJF1 A0A1B6CQX7 A0A182RBL4 Q7Q3L1 A0A182W9N1 A0A182M0V6 A0A182WT78 A0A1S4GWR5 A0A182HV73 A0A182G7K8 A0A182L217 A0A182Q4C0 A0A182THU0 A0A182K665 A0A182V637 A0A1Y1JZU7 A0A182YGU0 A0A1Y1K2U9 B3NLE4 A0A182F4L1 A0A182YGU1 A0A1Y1JZV1 A0A224XHL9 B4P8T4 A0A182PJ76 A0A182H6X9 A0A0J9R3R3 A0A182MNW7 Q7Q3L0 Q8SY13 A0A0Q5VUM4 A0A0R1DNZ4 A0A0A1XKB7 A0A2P8Z7Y1 W8B0U9 B4MTM9 B3MMJ5 A0A182PJ75 A0A1S4F2H1 A0A0K8UDR0 A0A3B0KAP3 Q17HG6 A0A182QG80 A0A1A9VL65 A0A3B0K2K1 A0A0P9BMX5 A0A034V4K5 A0A182W9N2 A0A1A9WUN8 B0X9B9 W8BCG7 A0A1I8P4K3 A0A0R3NUA0 Q29L84 A0A0K8VBJ8 A0A1B0A449 A0A182GT17 A0A182WT77 A0A182HV72 Q17HG5 B4JD11 A0A139WK34 A0A1I8P4R7 A0A1S4F2K6 A0A1S4GWQ7 K7J6Q0 A0A1B0AWA0 B4MDF2 A0A1A9YBP9
PDB
4OF8
E-value=1.25123e-05,
Score=115
Ontologies
GO
Topology
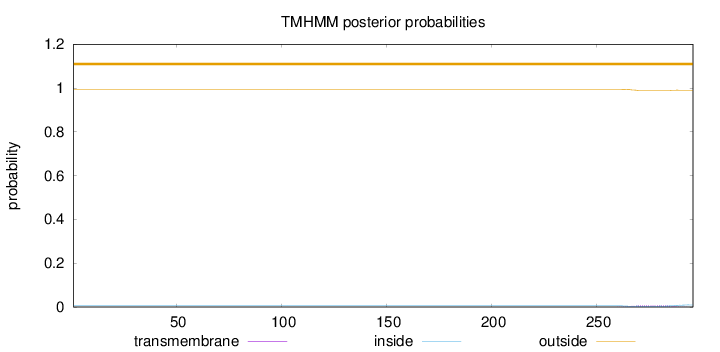
Length:
296
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.16465
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00561
outside
1 - 296
Population Genetic Test Statistics
Pi
24.645725
Theta
20.33243
Tajima's D
-1.393771
CLR
0.188958
CSRT
0.0750462476876156
Interpretation
Uncertain
