Gene
KWMTBOMO07406
Pre Gene Modal
BGIBMGA010344
Annotation
PREDICTED:_beta-1-syntrophin_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.193
Sequence
CDS
ATGGTGGAAAACGGTGCCTCGAGTGGTGGCAGTGATAGTCCTAGTGCAACATGTGTTCGTTCTGGTCTTCTAGAGACGTTAGTCCGTGGTGTTTGGTATCGCGTTCATTGTTCATTAGAAGACGATTACCTGAGCGTCTGTCTCGATGATGGGTATGATGCGACAACGACTTTGAATGGCACTCTGAACAACAACAACGTCGAAACTCCGAACAGCGACTTCGCAGAGGTACCCGAAGCAATTGCAAACCAAAAACGTTTAGTACAAGTTGTGAAATCGGACAACAACGGGCTTGGTATATCAATAAAAGGTGGAATCGAAAATAACATGCCAATATTGATATCAAAAATATTCAAGGGTATGGCAGCCGACTTGACTGAACAGTTGTACGTTGGTGATGCTATATTGTCTGTAAACGGTGAGGATCTCAAAGATGCGACACACGAAGAAGCTGTAAAGGCATTGAAGAGAGCTGGAAAAATGGTACAACTGGAAGTGAAATATCTCCGTGAAGTGACACCGTACTTCCGTAAGGCATCCATTATAAGTGAAGTAGGATGGGAGCTACAACGTGGGTATATGGCAGACACACCACCGTCGCCACCATCTCCACGCCGTCGACGGGCAGACACTCGTTATGTACCACTGTTAATGGCATGCGTTGCCAAGAATCTCCGTCATAATGACCCAGAGGATCGCACAATCGAAATATATTCACCGGACGGCGTCCACGCATTAGCATTACGGGCCAGTTCAGCGGCCACAGCGTCAGGGTGGCATCGCGCGCTACATGCAGCCGCACGTCGGGCTGCACGCGCCGCTCTTGCTCGCGCCAGGCCGAGACTTCGCCCTCTTGTAGGCGATGTGCGATATGCTGGTTGGATGGCAAGGCGCCCACCGCAAGATCATATGAGCGCTTCAGGAGGGTCTGACAGCTCCGACGAGACTGAAGGCTGGCAGCCAACATTCGTTGCAATCACAGACAGAGAGTTAAGGCTGTATGAGGCGGCGCCGTGGTCGGCGGAGGCATGGTGCGCGCCGGCGGAGAGCCTGCGGCTGGCGGCGACGCGGCTGGCCTGGTGGCGGCGAGCTGGGGCGGGCGGGGCGGCGGCGCTGGGGCTGCGGGCCGGGTGCGTGGACGGGCTGGTGCAGCGAGCCCTGCGCGCGGACTCGCCGCACGACCTGGCGGCCGCCGCGTCCGCGCTGGTGGAGGGGGCGCACGCGGCGGTGCGAGCGCAGCCCGAGTTCACGTTCCGCTGCTCGGTGGGCGGGGCGGCGGCGCGGCTGGCGCTGGGGGCGGCGGGCGTGTGCGTGTGGGAGGCGGCGGGCTCGCTGGGCCGCGCGCTGTACCGCCGCCCGCTGCACGCACTCCGCGCTTCCGCTGACGACGACAACAACAAGCTCTGGCTGCACTTCGCTGACGACGACACGTTGGAGCTAGAAATGGACGGTAGCCCGAAGCCGGCTGTGTTCATTCTGCACAATTTGCTGTCGGCTCGCGTCCACTCGCTGCCGGGAGATCCGACACCGAACCCGCTATAA
Protein
MVENGASSGGSDSPSATCVRSGLLETLVRGVWYRVHCSLEDDYLSVCLDDGYDATTTLNGTLNNNNVETPNSDFAEVPEAIANQKRLVQVVKSDNNGLGISIKGGIENNMPILISKIFKGMAADLTEQLYVGDAILSVNGEDLKDATHEEAVKALKRAGKMVQLEVKYLREVTPYFRKASIISEVGWELQRGYMADTPPSPPSPRRRRADTRYVPLLMACVAKNLRHNDPEDRTIEIYSPDGVHALALRASSAATASGWHRALHAAARRAARAALARARPRLRPLVGDVRYAGWMARRPPQDHMSASGGSDSSDETEGWQPTFVAITDRELRLYEAAPWSAEAWCAPAESLRLAATRLAWWRRAGAGGAAALGLRAGCVDGLVQRALRADSPHDLAAAASALVEGAHAAVRAQPEFTFRCSVGGAAARLALGAAGVCVWEAAGSLGRALYRRPLHALRASADDDNNKLWLHFADDDTLELEMDGSPKPAVFILHNLLSARVHSLPGDPTPNPL
Summary
Uniprot
H9JLE3
A0A212FLF6
A0A194Q051
A0A1B6JYL0
A0A1B6HJZ0
A0A1W4X6E1
+ More
A0A067R5A8 A0A0A9YGU7 A0A0V0G9Q3 A0A069DVA4 A0A023F4T2 T1HGP8 A0A1L8E3G4 A0A1L8E3S9 A0A3L8DK71 A0A0C9QT18 A0A2C9GQ69 K7IM22 A0A2M4CU94 Q7Q0A5 A0A084VI78 A0A1I8MAX9 A0A1I8MAY1 A0A1I8P747 A0A0A1WDQ1 A0A0K8V152 W8BKF2 A0A0C9R7Q3 B4LD77 A0A0M5IZA3 A0A0C9RNZ2 B4IYV9 A0A0R1E820 B4KUV6 B3MAT0 A0A0R1DYX2 B4QKM6 M9PGE3 Q9VNY4 B4IAV6 B3NIZ4 A0A0P9C2N2 B4PD37 A0A0J9UR75 Q9VNY2 A0A0Q5UKK4 A0A1W4URP9 A0A0J9RZQ8 A0A1W4URF7 A0A3B0JYI0 B4GRV4 Q2M0B5 A0A1D2MWG6
A0A067R5A8 A0A0A9YGU7 A0A0V0G9Q3 A0A069DVA4 A0A023F4T2 T1HGP8 A0A1L8E3G4 A0A1L8E3S9 A0A3L8DK71 A0A0C9QT18 A0A2C9GQ69 K7IM22 A0A2M4CU94 Q7Q0A5 A0A084VI78 A0A1I8MAX9 A0A1I8MAY1 A0A1I8P747 A0A0A1WDQ1 A0A0K8V152 W8BKF2 A0A0C9R7Q3 B4LD77 A0A0M5IZA3 A0A0C9RNZ2 B4IYV9 A0A0R1E820 B4KUV6 B3MAT0 A0A0R1DYX2 B4QKM6 M9PGE3 Q9VNY4 B4IAV6 B3NIZ4 A0A0P9C2N2 B4PD37 A0A0J9UR75 Q9VNY2 A0A0Q5UKK4 A0A1W4URP9 A0A0J9RZQ8 A0A1W4URF7 A0A3B0JYI0 B4GRV4 Q2M0B5 A0A1D2MWG6
Pubmed
EMBL
BABH01037166
AGBW02007778
OWR54571.1
KQ459592
KPI96795.1
GECU01003410
+ More
JAT04297.1 GECU01032719 JAS74987.1 KK852879 KDR14467.1 GBHO01011317 GDHC01019469 JAG32287.1 JAP99159.1 GECL01001355 JAP04769.1 GBGD01001257 JAC87632.1 GBBI01002723 JAC15989.1 ACPB03006856 GFDF01000816 JAV13268.1 GFDF01000815 JAV13269.1 QOIP01000007 RLU20807.1 GBYB01006879 JAG76646.1 APCN01003298 APCN01003299 GGFL01004573 MBW68751.1 AAAB01008986 EAA00191.4 ATLV01013325 ATLV01013326 ATLV01013327 ATLV01013328 ATLV01013329 KE524853 KFB37672.1 GBXI01017163 JAC97128.1 GDHF01019657 JAI32657.1 GAMC01004755 JAC01801.1 GBYB01008862 JAG78629.1 CH940647 EDW70984.1 CP012525 ALC45236.1 GBYB01008861 JAG78628.1 CH916366 EDV96646.1 CH891690 KRK05167.1 CH933809 EDW19362.1 CH902618 EDV39164.1 CM000159 KRK02169.1 CM000363 EDX11441.1 AE014296 AGB94884.1 AAF51779.3 CH480826 EDW44419.1 CH954178 EDV52711.1 KPU77922.1 EDW94969.2 KRK02168.1 CM002912 KMZ01091.1 BT024428 AAF51781.3 ABC86490.1 AGB94885.1 KQS44393.1 KQS44394.1 KMZ01092.1 KMZ01093.1 KMZ01094.1 OUUW01000002 SPP76158.1 CH479188 EDW40489.1 CH379069 EAL31020.2 LJIJ01000447 ODM97400.1
JAT04297.1 GECU01032719 JAS74987.1 KK852879 KDR14467.1 GBHO01011317 GDHC01019469 JAG32287.1 JAP99159.1 GECL01001355 JAP04769.1 GBGD01001257 JAC87632.1 GBBI01002723 JAC15989.1 ACPB03006856 GFDF01000816 JAV13268.1 GFDF01000815 JAV13269.1 QOIP01000007 RLU20807.1 GBYB01006879 JAG76646.1 APCN01003298 APCN01003299 GGFL01004573 MBW68751.1 AAAB01008986 EAA00191.4 ATLV01013325 ATLV01013326 ATLV01013327 ATLV01013328 ATLV01013329 KE524853 KFB37672.1 GBXI01017163 JAC97128.1 GDHF01019657 JAI32657.1 GAMC01004755 JAC01801.1 GBYB01008862 JAG78629.1 CH940647 EDW70984.1 CP012525 ALC45236.1 GBYB01008861 JAG78628.1 CH916366 EDV96646.1 CH891690 KRK05167.1 CH933809 EDW19362.1 CH902618 EDV39164.1 CM000159 KRK02169.1 CM000363 EDX11441.1 AE014296 AGB94884.1 AAF51779.3 CH480826 EDW44419.1 CH954178 EDV52711.1 KPU77922.1 EDW94969.2 KRK02168.1 CM002912 KMZ01091.1 BT024428 AAF51781.3 ABC86490.1 AGB94885.1 KQS44393.1 KQS44394.1 KMZ01092.1 KMZ01093.1 KMZ01094.1 OUUW01000002 SPP76158.1 CH479188 EDW40489.1 CH379069 EAL31020.2 LJIJ01000447 ODM97400.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000192223
UP000027135
UP000015103
+ More
UP000279307 UP000075840 UP000002358 UP000007062 UP000030765 UP000095301 UP000095300 UP000008792 UP000092553 UP000001070 UP000002282 UP000009192 UP000007801 UP000000304 UP000000803 UP000001292 UP000008711 UP000192221 UP000268350 UP000008744 UP000001819 UP000094527
UP000279307 UP000075840 UP000002358 UP000007062 UP000030765 UP000095301 UP000095300 UP000008792 UP000092553 UP000001070 UP000002282 UP000009192 UP000007801 UP000000304 UP000000803 UP000001292 UP000008711 UP000192221 UP000268350 UP000008744 UP000001819 UP000094527
Interpro
SUPFAM
SSF50156
SSF50156
Gene 3D
ProteinModelPortal
H9JLE3
A0A212FLF6
A0A194Q051
A0A1B6JYL0
A0A1B6HJZ0
A0A1W4X6E1
+ More
A0A067R5A8 A0A0A9YGU7 A0A0V0G9Q3 A0A069DVA4 A0A023F4T2 T1HGP8 A0A1L8E3G4 A0A1L8E3S9 A0A3L8DK71 A0A0C9QT18 A0A2C9GQ69 K7IM22 A0A2M4CU94 Q7Q0A5 A0A084VI78 A0A1I8MAX9 A0A1I8MAY1 A0A1I8P747 A0A0A1WDQ1 A0A0K8V152 W8BKF2 A0A0C9R7Q3 B4LD77 A0A0M5IZA3 A0A0C9RNZ2 B4IYV9 A0A0R1E820 B4KUV6 B3MAT0 A0A0R1DYX2 B4QKM6 M9PGE3 Q9VNY4 B4IAV6 B3NIZ4 A0A0P9C2N2 B4PD37 A0A0J9UR75 Q9VNY2 A0A0Q5UKK4 A0A1W4URP9 A0A0J9RZQ8 A0A1W4URF7 A0A3B0JYI0 B4GRV4 Q2M0B5 A0A1D2MWG6
A0A067R5A8 A0A0A9YGU7 A0A0V0G9Q3 A0A069DVA4 A0A023F4T2 T1HGP8 A0A1L8E3G4 A0A1L8E3S9 A0A3L8DK71 A0A0C9QT18 A0A2C9GQ69 K7IM22 A0A2M4CU94 Q7Q0A5 A0A084VI78 A0A1I8MAX9 A0A1I8MAY1 A0A1I8P747 A0A0A1WDQ1 A0A0K8V152 W8BKF2 A0A0C9R7Q3 B4LD77 A0A0M5IZA3 A0A0C9RNZ2 B4IYV9 A0A0R1E820 B4KUV6 B3MAT0 A0A0R1DYX2 B4QKM6 M9PGE3 Q9VNY4 B4IAV6 B3NIZ4 A0A0P9C2N2 B4PD37 A0A0J9UR75 Q9VNY2 A0A0Q5UKK4 A0A1W4URP9 A0A0J9RZQ8 A0A1W4URF7 A0A3B0JYI0 B4GRV4 Q2M0B5 A0A1D2MWG6
PDB
1Z87
E-value=3.13322e-40,
Score=416
Ontologies
GO
PANTHER
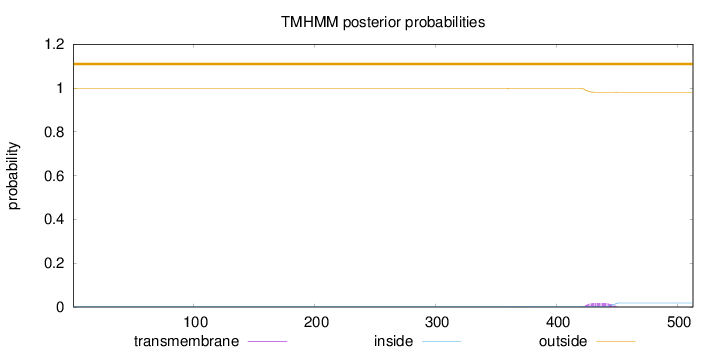
Topology
Length:
513
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.408
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00100
outside
1 - 513
Population Genetic Test Statistics
Pi
23.279917
Theta
24.741919
Tajima's D
-0.692184
CLR
0.931157
CSRT
0.198790060496975
Interpretation
Uncertain
