Gene
KWMTBOMO07402
Pre Gene Modal
BGIBMGA010343
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_tyrosine-protein_kinase_transmembrane_receptor_Ror-like_[Bombyx_mori]
Full name
Tyrosine-protein kinase receptor
Location in the cell
Nuclear Reliability : 2.567
Sequence
CDS
ATGAAATGTATTACAGTGTTAGTTATGTGCCTGTGTTTACATGACATTCATTCATCAAGTGAAGAGAAAACAGAATCTATCTCTCCTCTAAGACTTAATAGTACATCAGAATGGAACATCAAACTGGATATATTCTTTTCTAAATCTTTACCTCAGCATGTAACGAGATCTCCTGGTAGTGATGTTCGTTTTAAATGTGAAGCTTACATAAACTTTACGAAAGTTGAAAAGTACACTAAAAATGAACAAGGTATAGAAACTTTACATGCTGAGCTACCCCCGCAGCATATACAGCAAAGACTGTTGAACAATGTAAAAGTATATTGGCTCAAAAACAATAAAAGTATTGAAATTTCAAACAAAGTTAATGTTACAACTAAAATCAATAATATAACGATTTCCACAAATTTAAAGATCAGAAACATAAGTAATGGTGATGCAGGTAACTACACATGTGTTTACAAACAAATGTATGAAAAAAAGCAAACTTCCTTACTCAAAGTTGAAGATAGTGTTGACATACCTACCCATGCACCATATTTCCCAAATTCAAATTTTGTAAATGAATTAAGAATTACAACCCCGGATGTGGTGTCAGAAAATATTAAAACTATGAATATAAACGTTACTGATTCTCCTAATATACCTCTTCAAAAACTTGAACCAATTTGTCAAGAATATGTTGGGAAAGTTTGTGCATTTCACTTAAGAGGCCAATCTGTTTATATCCCATATGATACCACACAGATTGAATTGGAAGAGAAGTTAAGAAAAGCAATACAAGTTACAAGATTTTCTAATGAAATAAGTTCACACTGTGAAAGATATGCATTACCTAGTTTGTGCTACTCCACTTTTCCCATATGTAGAAATCCTGTAGCTACAAATGCTAATTTCTTTAAAGATGCAAGACAGGTATTTCTCATATCAAATAGAACCAACACTGCAGATTTTCCTGAAGTTGTATTTGAAAATATGCCTAATGGAAATGTACCTAATACTTTTTACTTGAAACCAAACAATATATCAAATGAAATGTTTAACTATCGTCATAATACAACAATACTGAGGAGAGTATGTAAGAAAGAGTGTGAAATTTTAGAAACAGAATTATGTCAGAAAGAATATGCCATAGCGAAAAGGCATCCCCATATTGGTCAGCAATTAACTTTAGAAGAATGTCAAGATTTACCTGAAAATGATCCAGACTGTCTTAAAATTGAAATTGGAGCTTTAATGATTTCAGATGATGAGTGTTATTGGGAAAACGGCAGTAGTTATTTGGGCAAAGTTAATGTTGCCAGTAATGGGTTGGCTTGTATTGAATGGTCAAAGCAAATGTATGTTACATTATCAAATTACCCTGTACTTGCTGGTAGACACAGCTACTGCCGTAACCCAGGAGGAGTCAATGAACAGCCTTGGTGTGTGGTAGATAAAGATGGTAAACTATATAAACAATTTTGTAACATTCCCAAATGTGCTCACAAACTTTGGATATACATAGTTGTTATCTTGGCAGTATCGTTCATAGTTATTGTGGGATTTGTTGTTTGTCTCATTTGCAGACATAAAAAAAAGAGTAATGCTGCTGCAATTAGAGACATTAACTTACCTAATGCTGATAAGAATATCTATGGAAATTCTAGATTGAATTCTCCAATAGAAATGACAGACTTACTGCCAAACCATAGTAATCAGCCCCATTTGACCAGGAGTACTCGTGGAAATGGTGTACTCAGAATACCTCAATATTCACTGTCGCAGATCAAATTCCTGGAAGAGTTAGGGGAGGGAGCTTTTGGAAAAGTTTACAAAGGAGCACTGAAAAAGAGTGGAGATACACAGTTTATAGCAGTCAAAGCACTCAAAGAAAATGCTTCAGCTAAGACCAAGGCTGATTTCAGAAGAGAAATTGATTTGATTTCTGAACTTAACCATGACAATATTGTGTGTATTGTTGGTGTTGCTTTAAGTGAAGAACCACTTTGCATGTTATTTGAGTTCATGGCTCGAGGAGACTTGCATGAATTTTTAATGGGAAGAGCACCTCCATCAGGAAAGGGTTTACCTCCCATGAGACTGCTGAACATTGCACATAATATTGCATCAGGCATGCAGTATTTGGCCTCACACCATTATGTACATAGAGATTTAGCTGCAAGAAATTGTTTGGTTTCCGATGATTTTGTTGTGAAGATATCTGATTTTGGCCTCTCTAGAGATATATATAGTTCAGATTATTACAGAGTACAATCAAAAAGTTTATTGCCCGTTAGATGGATGCCACCAGAATCTATACTTTATGGAAAATTCACTACTGAAAGTGATATATGGTCATATGGGGTTGTCCTGTGGGAAATTTACAGTTATGGCCTCCAGCCTTATTATGGTTATAGTAATCAAGAAGTCATATCGATGGTGCGGAGTGGTGAGTTTTTAGCAGCACCATCAGCATGTCCATTGCCCATGTATGAACTCATGAGGGACTGTTGGAAACACACACCACATCGAAGGCCATCATTTGAAGATATAGTTAACAGAATTCAAGAGTGGATCCAGATGGGGGGATGTTCAGATGTTGGACCATCAGAATCTAGCAGCAATATAAGCGCGTACAGAACGACTAGTACTAGCAGGGATCTTCAGGAAAGAGTTCCACTGTTGCCCCCTCATTGCTCTTCATCTAATGGCTCTCTTGCCACCGGGAGCTTAAAGAAATTTAGTGTAGCTGGTTCAAGTTCGAAGGTAACTGATGATAATTGCTCTACATGTAGTGAAGTACCCCCATTAGCTCCAAAATCTAAAAAACACAGTGCAGCATCGCTGATAGACTCAGACAAAATAGGCAACATGAGTACAAGGGAGACAATCATTAGAATACCTAATGCACAGCACGGTAATGAAATTTAG
Protein
MKCITVLVMCLCLHDIHSSSEEKTESISPLRLNSTSEWNIKLDIFFSKSLPQHVTRSPGSDVRFKCEAYINFTKVEKYTKNEQGIETLHAELPPQHIQQRLLNNVKVYWLKNNKSIEISNKVNVTTKINNITISTNLKIRNISNGDAGNYTCVYKQMYEKKQTSLLKVEDSVDIPTHAPYFPNSNFVNELRITTPDVVSENIKTMNINVTDSPNIPLQKLEPICQEYVGKVCAFHLRGQSVYIPYDTTQIELEEKLRKAIQVTRFSNEISSHCERYALPSLCYSTFPICRNPVATNANFFKDARQVFLISNRTNTADFPEVVFENMPNGNVPNTFYLKPNNISNEMFNYRHNTTILRRVCKKECEILETELCQKEYAIAKRHPHIGQQLTLEECQDLPENDPDCLKIEIGALMISDDECYWENGSSYLGKVNVASNGLACIEWSKQMYVTLSNYPVLAGRHSYCRNPGGVNEQPWCVVDKDGKLYKQFCNIPKCAHKLWIYIVVILAVSFIVIVGFVVCLICRHKKKSNAAAIRDINLPNADKNIYGNSRLNSPIEMTDLLPNHSNQPHLTRSTRGNGVLRIPQYSLSQIKFLEELGEGAFGKVYKGALKKSGDTQFIAVKALKENASAKTKADFRREIDLISELNHDNIVCIVGVALSEEPLCMLFEFMARGDLHEFLMGRAPPSGKGLPPMRLLNIAHNIASGMQYLASHHYVHRDLAARNCLVSDDFVVKISDFGLSRDIYSSDYYRVQSKSLLPVRWMPPESILYGKFTTESDIWSYGVVLWEIYSYGLQPYYGYSNQEVISMVRSGEFLAAPSACPLPMYELMRDCWKHTPHRRPSFEDIVNRIQEWIQMGGCSDVGPSESSSNISAYRTTSTSRDLQERVPLLPPHCSSSNGSLATGSLKKFSVAGSSSKVTDDNCSTCSEVPPLAPKSKKHSAASLIDSDKIGNMSTRETIIRIPNAQHGNEI
Summary
Catalytic Activity
ATP + L-tyrosyl-[protein] = ADP + H(+) + O-phospho-L-tyrosyl-[protein]
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family. Insulin receptor subfamily.
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Feature
chain Tyrosine-protein kinase receptor
Uniprot
A0A2A4K4D3
A0A3S2THF9
A0A194R429
A0A194PUX8
A0A0L7L2X6
A0A1S4FQ29
+ More
W8C503 A0A336MEA8 A0A336LGT4 A0A3L8DZ56 A0A195EY13 A0A151WKE8 A0A195CE65 A0A195BQD8 A0A154P7U0 A0A0J7KNY4 E9IQD9 E2C8Q0 F4W9T8 A0A0M9A8C4 A0A158NYX7 A0A088A947 D2A6A0 E2A5T3 A0A0L7R2I1 A0A2A3E0N0 A0A232FIZ5 K7J9R8 A0A026W8W6 A0A151JB73 A0A1Y1M7L8 A0A1Y1M0D5 A0A1Y1M7M7 A0A1Y1M5Z5 A0A3B0K6J9 A0A0C9R3F9 A0A336K6V3 T1HCV4 A0A146M6A3 A0A0A9Z5Y9 A0A0U3K0U7 A0A1B6J519 T1IJ71 A0A1B6LIV8
W8C503 A0A336MEA8 A0A336LGT4 A0A3L8DZ56 A0A195EY13 A0A151WKE8 A0A195CE65 A0A195BQD8 A0A154P7U0 A0A0J7KNY4 E9IQD9 E2C8Q0 F4W9T8 A0A0M9A8C4 A0A158NYX7 A0A088A947 D2A6A0 E2A5T3 A0A0L7R2I1 A0A2A3E0N0 A0A232FIZ5 K7J9R8 A0A026W8W6 A0A151JB73 A0A1Y1M7L8 A0A1Y1M0D5 A0A1Y1M7M7 A0A1Y1M5Z5 A0A3B0K6J9 A0A0C9R3F9 A0A336K6V3 T1HCV4 A0A146M6A3 A0A0A9Z5Y9 A0A0U3K0U7 A0A1B6J519 T1IJ71 A0A1B6LIV8
EC Number
2.7.10.1
Pubmed
EMBL
NWSH01000173
PCG78768.1
RSAL01000139
RVE46142.1
KQ460845
KPJ12000.1
+ More
KQ459592 KPI96793.1 JTDY01003386 KOB69634.1 GAMC01009286 JAB97269.1 UFQT01001062 SSX28694.1 UFQS01000002 UFQT01000002 SSW96699.1 SSX17086.1 QOIP01000002 RLU25724.1 KQ981909 KYN33175.1 KQ983012 KYQ48342.1 KQ977876 KYM99097.1 KQ976423 KYM88786.1 KQ434839 KZC07989.1 LBMM01004843 KMQ92042.1 GL764736 EFZ17209.1 GL453671 EFN75674.1 GL888033 EGI69074.1 KQ435721 KOX78546.1 ADTU01000806 KQ971346 EFA05483.2 GL437059 EFN71205.1 KQ414666 KOC65085.1 KZ288477 PBC25280.1 NNAY01000158 OXU30449.1 AAZX01001946 KK107329 EZA52500.1 KQ979229 KYN22205.1 GEZM01043121 JAV79307.1 GEZM01043124 JAV79302.1 GEZM01043117 GEZM01043113 JAV79317.1 GEZM01043116 GEZM01043114 JAV79326.1 OUUW01000006 SPP81639.1 GBYB01007322 JAG77089.1 SSW96700.1 SSX17087.1 ACPB03000222 ACPB03000223 GDHC01003328 JAQ15301.1 GBHO01004304 GBHO01004303 JAG39300.1 JAG39301.1 KT355588 ALV82507.1 GECU01013434 JAS94272.1 JH430232 GEBQ01016378 JAT23599.1
KQ459592 KPI96793.1 JTDY01003386 KOB69634.1 GAMC01009286 JAB97269.1 UFQT01001062 SSX28694.1 UFQS01000002 UFQT01000002 SSW96699.1 SSX17086.1 QOIP01000002 RLU25724.1 KQ981909 KYN33175.1 KQ983012 KYQ48342.1 KQ977876 KYM99097.1 KQ976423 KYM88786.1 KQ434839 KZC07989.1 LBMM01004843 KMQ92042.1 GL764736 EFZ17209.1 GL453671 EFN75674.1 GL888033 EGI69074.1 KQ435721 KOX78546.1 ADTU01000806 KQ971346 EFA05483.2 GL437059 EFN71205.1 KQ414666 KOC65085.1 KZ288477 PBC25280.1 NNAY01000158 OXU30449.1 AAZX01001946 KK107329 EZA52500.1 KQ979229 KYN22205.1 GEZM01043121 JAV79307.1 GEZM01043124 JAV79302.1 GEZM01043117 GEZM01043113 JAV79317.1 GEZM01043116 GEZM01043114 JAV79326.1 OUUW01000006 SPP81639.1 GBYB01007322 JAG77089.1 SSW96700.1 SSX17087.1 ACPB03000222 ACPB03000223 GDHC01003328 JAQ15301.1 GBHO01004304 GBHO01004303 JAG39300.1 JAG39301.1 KT355588 ALV82507.1 GECU01013434 JAS94272.1 JH430232 GEBQ01016378 JAT23599.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000053268
UP000037510
UP000279307
+ More
UP000078541 UP000075809 UP000078542 UP000078540 UP000076502 UP000036403 UP000008237 UP000007755 UP000053105 UP000005205 UP000005203 UP000007266 UP000000311 UP000053825 UP000242457 UP000215335 UP000002358 UP000053097 UP000078492 UP000268350 UP000015103
UP000078541 UP000075809 UP000078542 UP000078540 UP000076502 UP000036403 UP000008237 UP000007755 UP000053105 UP000005205 UP000005203 UP000007266 UP000000311 UP000053825 UP000242457 UP000215335 UP000002358 UP000053097 UP000078492 UP000268350 UP000015103
Interpro
IPR000719
Prot_kinase_dom
+ More
IPR036179 Ig-like_dom_sf
IPR008266 Tyr_kinase_AS
IPR002011 Tyr_kinase_rcpt_2_CS
IPR020635 Tyr_kinase_cat_dom
IPR013783 Ig-like_fold
IPR017441 Protein_kinase_ATP_BS
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR018056 Kringle_CS
IPR020067 Frizzled_dom
IPR041775 Ror-like_CRD
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR013098 Ig_I-set
IPR011009 Kinase-like_dom_sf
IPR038178 Kringle_sf
IPR000001 Kringle
IPR013806 Kringle-like
IPR036790 Frizzled_dom_sf
IPR003598 Ig_sub2
IPR013151 Immunoglobulin
IPR036179 Ig-like_dom_sf
IPR008266 Tyr_kinase_AS
IPR002011 Tyr_kinase_rcpt_2_CS
IPR020635 Tyr_kinase_cat_dom
IPR013783 Ig-like_fold
IPR017441 Protein_kinase_ATP_BS
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR018056 Kringle_CS
IPR020067 Frizzled_dom
IPR041775 Ror-like_CRD
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR013098 Ig_I-set
IPR011009 Kinase-like_dom_sf
IPR038178 Kringle_sf
IPR000001 Kringle
IPR013806 Kringle-like
IPR036790 Frizzled_dom_sf
IPR003598 Ig_sub2
IPR013151 Immunoglobulin
Gene 3D
ProteinModelPortal
A0A2A4K4D3
A0A3S2THF9
A0A194R429
A0A194PUX8
A0A0L7L2X6
A0A1S4FQ29
+ More
W8C503 A0A336MEA8 A0A336LGT4 A0A3L8DZ56 A0A195EY13 A0A151WKE8 A0A195CE65 A0A195BQD8 A0A154P7U0 A0A0J7KNY4 E9IQD9 E2C8Q0 F4W9T8 A0A0M9A8C4 A0A158NYX7 A0A088A947 D2A6A0 E2A5T3 A0A0L7R2I1 A0A2A3E0N0 A0A232FIZ5 K7J9R8 A0A026W8W6 A0A151JB73 A0A1Y1M7L8 A0A1Y1M0D5 A0A1Y1M7M7 A0A1Y1M5Z5 A0A3B0K6J9 A0A0C9R3F9 A0A336K6V3 T1HCV4 A0A146M6A3 A0A0A9Z5Y9 A0A0U3K0U7 A0A1B6J519 T1IJ71 A0A1B6LIV8
W8C503 A0A336MEA8 A0A336LGT4 A0A3L8DZ56 A0A195EY13 A0A151WKE8 A0A195CE65 A0A195BQD8 A0A154P7U0 A0A0J7KNY4 E9IQD9 E2C8Q0 F4W9T8 A0A0M9A8C4 A0A158NYX7 A0A088A947 D2A6A0 E2A5T3 A0A0L7R2I1 A0A2A3E0N0 A0A232FIZ5 K7J9R8 A0A026W8W6 A0A151JB73 A0A1Y1M7L8 A0A1Y1M0D5 A0A1Y1M7M7 A0A1Y1M5Z5 A0A3B0K6J9 A0A0C9R3F9 A0A336K6V3 T1HCV4 A0A146M6A3 A0A0A9Z5Y9 A0A0U3K0U7 A0A1B6J519 T1IJ71 A0A1B6LIV8
Ontologies
GO
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.992222
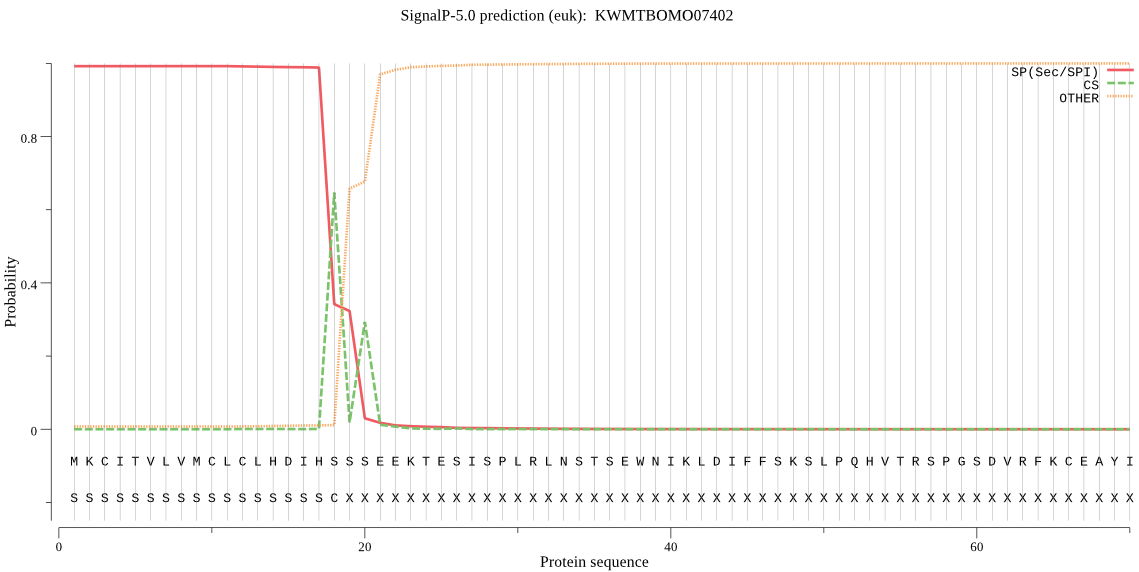
Length:
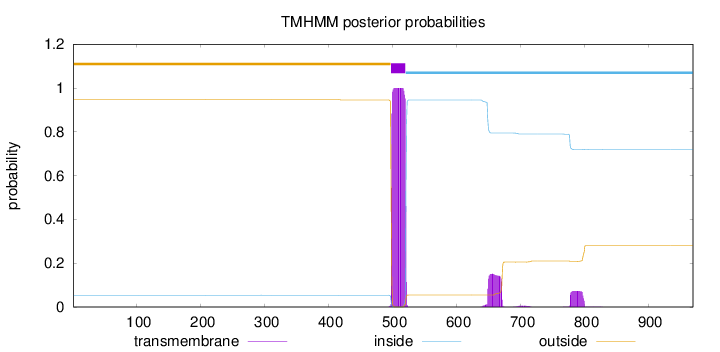
970
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
27.92171
Exp number, first 60 AAs:
0.00016
Total prob of N-in:
0.05399
outside
1 - 497
TMhelix
498 - 520
inside
521 - 970
Population Genetic Test Statistics
Pi
17.735233
Theta
14.965264
Tajima's D
0.668771
CLR
0.478898
CSRT
0.568921553922304
Interpretation
Uncertain
