Gene
KWMTBOMO07401
Pre Gene Modal
BGIBMGA010322
Annotation
Leucine-rich_repeat-containing_protein_40_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.564 Nuclear Reliability : 2.019
Sequence
CDS
ATGGGCTTGGTAAAATCAGCGAAGAGAACTGGACAGTTAAGTTTGTGCAACAGAGGTCTTGGAACAGTACCAGAAAATGTTTGGAAGATAGATGAATTAGTAATGGATGAAACTAAAGATGTAGATTTTACAAGATCAGATCAACAGAGTTGGTGGAATTATGAGCCATTAAAGACTCTGGATCTCAGTTCAAATGTTATAAAAATTATATCTCCGAATGTGAGACTTTTACAACAACTTATTACACTCAAGTTGCACGACAACGCTATTGAAAGCTTACCACCGGAATTTGGTGACCTCAAGAATTTATCGAATTTAAGCTTAGACCACAATCACCTGTCAGTTCTACCTAAAGAGTTCTACAGATTAACGGAACTGAGGTGGCTCGGTCTATCACACAATGCACTGAAAAAAATCGAACCAGATTTCGGAGATCTTGTCATGTTAAATTTTTTAGACTTGTCACACAACAAACTAACAGTATTACCTCCCGGAATGGGATATCTCGTGCGTCTAGTCGAACTGAATCTTTCGCACAACCAGCTACTTGAGCTACCGCCTGATATTGTCAATTTAAGAGATTTGAAAAAGCTAAACATTAGTAACAATAATCTAAAAAAGATGCCTCCGCTAGGCGAGTTGAGAAAAATGGAAATTTTGGACGCAAACCATAATGATATTGAAGAGTTGCCAGACTTCTACGGTTGTACGGCGCTCAAGGAAATATATTTAGCCAACAATTATATCAAAGAGATAACCGAAGAGTTTTGCGACCAAATGCAACACTTGAGCATATTAAACATACGCGACAATAAACTCGAAGTGTTGCCAGAGAACATATCACTATTGCAAAAATTGAAAAGGCTGGACCTAACTAATAACAATTTAAATAAGTTGCCAAGAAATTTAGGTTTAATGTCACAATTACAAAGCATCAACATGGACGGTAACAGGCTGTCGTTCGTGAGGCAGGACATCATCAGAGGAGGCACCGATCGCATGATGAAGTTTCTTCGAGATCGTATCACGGAGGAAATTTTAGCTGAAACCAGACTGGAAAACGAGTGGCCAGACAAATACACGTTGAAGAAAAGTCAGGCGCTGATGTTGCCATCGCGCGAGCTACAAAACGTACCAGACGACGTGTTCAAAGCGGCAGCCGATGCGGAGGTGCATATTATAGATCTGTCCAAGAACAAGTTAACCAGTGTGCCTTTTGGAATCTGTCAAGTTAAAGAATCGCTGACGCAATTAGTGCTATCGTCGAACAATATATCGAATTTTCCACCGGAGATAAGCAACTGCGTACATATCCAGTATATAGACTTGGGGAAGAACTGTCTCACCGAATTACCAAAGGAGATTGGTTCCTTGAAATACTTAAGGGAACTCGTCATATCGAATAACAAGTTTACGAAAATCCCTCGTTGCGTTTATGATTTGGAGAACTTGGAGATATTGCTGGCCGCCGATAATCAGATCGAGGAAATTAATGTATCGTCGGACGCGCTCGCCCGGCTCGGCAAGCTGGCCGTCCTTGACCTGGCCAATAACAGCATCTCTACAGTACCGCCCGAGCTAGGGAACTTCACGCATCTGAGGACACTGGAGTTGATGGGCAACTGTTTCCGTCAGCCTCGGCATGCCGTGTTGGCGAAGGGCACCGCCTCAGTGCTCTCGTACCTGCGGGACCGGATCCCTACCGCCTAG
Protein
MGLVKSAKRTGQLSLCNRGLGTVPENVWKIDELVMDETKDVDFTRSDQQSWWNYEPLKTLDLSSNVIKIISPNVRLLQQLITLKLHDNAIESLPPEFGDLKNLSNLSLDHNHLSVLPKEFYRLTELRWLGLSHNALKKIEPDFGDLVMLNFLDLSHNKLTVLPPGMGYLVRLVELNLSHNQLLELPPDIVNLRDLKKLNISNNNLKKMPPLGELRKMEILDANHNDIEELPDFYGCTALKEIYLANNYIKEITEEFCDQMQHLSILNIRDNKLEVLPENISLLQKLKRLDLTNNNLNKLPRNLGLMSQLQSINMDGNRLSFVRQDIIRGGTDRMMKFLRDRITEEILAETRLENEWPDKYTLKKSQALMLPSRELQNVPDDVFKAAADAEVHIIDLSKNKLTSVPFGICQVKESLTQLVLSSNNISNFPPEISNCVHIQYIDLGKNCLTELPKEIGSLKYLRELVISNNKFTKIPRCVYDLENLEILLAADNQIEEINVSSDALARLGKLAVLDLANNSISTVPPELGNFTHLRTLELMGNCFRQPRHAVLAKGTASVLSYLRDRIPTA
Summary
Uniprot
H9JLC1
A0A2H1X2Z0
A0A2A4K533
A0A194PTQ0
A0A3S2NFE5
A0A212F3I0
+ More
A0A194R2S7 D7EI93 A0A1W4W897 A0A0K8TM45 A0A1Y1KX67 A0A067RXA1 A0A084WRP7 A0A182IS19 A0A182R9U5 A0A1Q3FF44 Q17FY2 A0A1S4F485 Q7QIP3 A0A182UVN1 A0A182Y318 A0A182P131 A0A182L3H7 A0A182I6V5 A0A232FJA9 A0A182TET1 A0A2M4AIC5 A0A1J1HL64 B0W215 A0A182WWL9 W5J893 A0A182QWH2 A0A2M3Z5X9 A0A2M3Z5Q3 A0A2M3Z5V8 T1PGZ9 A0A2M4BHS4 A0A2M4BHB7 A0A2M4BHB0 A0A182LZR7 A0A1I8MZJ6 K7J196 A0A1I8NUC0 U4U5Y7 A0A182S9M1 B4KBR1 A0A0L0BN41 A0A034VZG8 A0A0A1XGE0 A0A154PA54 A0A1I8MZJ8 A0A088AII6 A0A1I8NUB6 A0A0Q9VZN2 B3LWV9 A0A182WHX4 A0A0K8VA22 A0A1W4V588 A0A182FGB9 Q7KSK8 A0A0M4EUK2 A0A0N0BJ46 A0A0T6AVY2 A0A1B0CLB2 A0A0K8W2I9 A0A3B0JHX2 A0A1Y1KZS2 A0A0R1E7F9 Q9V3X1 A0A0C9QVY5 A0A034VZH2 A0A3B0K5Q2 B5DXD8 A0A310SU42 B4G3J2 B4JYB6 A0A1W4USX3 A0A1B0D8B2 A0A0Q9VZN4 A0A0P8ZZ49 A0A131XDE8 A0A0R1E6W6 A8JQZ7 A0A2A3EK13 Q86NW5 A0A3B0JD55 A0A151IGD6 E2A1S0 I5ANK6 A0A3L8DHT2 A0A0P4WRX5 A0A195BW80 V5HSX0 A0A158NCD3 A0A151WS46 A0A195F9E9 A0A131Y2L8 A0A151ITJ9 A0A023EUN6 L7MGG2
A0A194R2S7 D7EI93 A0A1W4W897 A0A0K8TM45 A0A1Y1KX67 A0A067RXA1 A0A084WRP7 A0A182IS19 A0A182R9U5 A0A1Q3FF44 Q17FY2 A0A1S4F485 Q7QIP3 A0A182UVN1 A0A182Y318 A0A182P131 A0A182L3H7 A0A182I6V5 A0A232FJA9 A0A182TET1 A0A2M4AIC5 A0A1J1HL64 B0W215 A0A182WWL9 W5J893 A0A182QWH2 A0A2M3Z5X9 A0A2M3Z5Q3 A0A2M3Z5V8 T1PGZ9 A0A2M4BHS4 A0A2M4BHB7 A0A2M4BHB0 A0A182LZR7 A0A1I8MZJ6 K7J196 A0A1I8NUC0 U4U5Y7 A0A182S9M1 B4KBR1 A0A0L0BN41 A0A034VZG8 A0A0A1XGE0 A0A154PA54 A0A1I8MZJ8 A0A088AII6 A0A1I8NUB6 A0A0Q9VZN2 B3LWV9 A0A182WHX4 A0A0K8VA22 A0A1W4V588 A0A182FGB9 Q7KSK8 A0A0M4EUK2 A0A0N0BJ46 A0A0T6AVY2 A0A1B0CLB2 A0A0K8W2I9 A0A3B0JHX2 A0A1Y1KZS2 A0A0R1E7F9 Q9V3X1 A0A0C9QVY5 A0A034VZH2 A0A3B0K5Q2 B5DXD8 A0A310SU42 B4G3J2 B4JYB6 A0A1W4USX3 A0A1B0D8B2 A0A0Q9VZN4 A0A0P8ZZ49 A0A131XDE8 A0A0R1E6W6 A8JQZ7 A0A2A3EK13 Q86NW5 A0A3B0JD55 A0A151IGD6 E2A1S0 I5ANK6 A0A3L8DHT2 A0A0P4WRX5 A0A195BW80 V5HSX0 A0A158NCD3 A0A151WS46 A0A195F9E9 A0A131Y2L8 A0A151ITJ9 A0A023EUN6 L7MGG2
Pubmed
19121390
26354079
22118469
18362917
19820115
26369729
+ More
28004739 24845553 24438588 17510324 12364791 14747013 17210077 25244985 20966253 28648823 20920257 23761445 25315136 20075255 23537049 17994087 26108605 25348373 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 28049606 20798317 30249741 25765539 21347285 24945155 25576852
28004739 24845553 24438588 17510324 12364791 14747013 17210077 25244985 20966253 28648823 20920257 23761445 25315136 20075255 23537049 17994087 26108605 25348373 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 28049606 20798317 30249741 25765539 21347285 24945155 25576852
EMBL
BABH01037159
BABH01037160
ODYU01013009
SOQ59612.1
NWSH01000173
PCG78770.1
+ More
KQ459592 KPI96791.1 RSAL01000139 RVE46138.1 AGBW02010564 OWR48298.1 KQ460845 KPJ12002.1 KQ972669 EFA11666.2 GDAI01002166 JAI15437.1 GEZM01071157 JAV65999.1 KK852405 KDR24519.1 ATLV01026175 KE525407 KFB52891.1 GFDL01008875 JAV26170.1 CH477267 EAT45492.1 AAAB01008807 EAA04662.4 APCN01003701 NNAY01000140 OXU30630.1 GGFK01007161 MBW40482.1 CVRI01000010 CRK88773.1 DS231824 EDS27451.1 ADMH02002070 ETN59613.1 AXCN02000985 GGFM01003097 MBW23848.1 GGFM01003092 MBW23843.1 GGFM01003152 MBW23903.1 KA648082 AFP62711.1 GGFJ01003423 MBW52564.1 GGFJ01003309 MBW52450.1 GGFJ01003308 MBW52449.1 AXCM01000491 KB631679 ERL85345.1 CH933806 EDW14738.1 JRES01001623 KNC21417.1 GAKP01011455 JAC47497.1 GBXI01004255 JAD10037.1 KQ434856 KZC08775.1 CH940656 KRF78316.1 CH902617 EDV42747.2 GDHF01016510 JAI35804.1 AF184226 AE014297 AAD55737.1 AAF54976.1 CP012526 ALC46792.1 KQ435720 KOX78634.1 LJIG01022741 KRT78958.1 AJWK01017069 GDHF01007043 JAI45271.1 OUUW01000005 SPP80313.1 GEZM01071155 JAV66001.1 CM000160 KRK03620.1 AAF54977.3 GBYB01004806 GBYB01004807 JAG74573.1 JAG74574.1 GAKP01011450 JAC47502.1 SPP80311.1 CM000070 EDY67874.2 KQ760444 OAD60283.1 CH479179 EDW25000.1 CH916377 EDV90678.1 AJVK01004027 AJVK01004028 AJVK01004029 KRF78313.1 KPU79889.1 GEFH01004461 JAP64120.1 KRK03617.1 ABW08664.1 KZ288222 PBC32038.1 BT003605 AAO39608.1 SPP80314.1 KQ977728 KYN00260.1 GL435766 EFN72743.1 EIM52541.2 QOIP01000008 RLU19977.1 GDRN01038292 JAI67260.1 KQ976396 KYM92889.1 GANP01007215 JAB77253.1 ADTU01011818 KQ982796 KYQ50617.1 KQ981727 KYN37068.1 GEFM01003048 JAP72748.1 KQ981004 KYN10391.1 GAPW01000916 JAC12682.1 GACK01002117 JAA62917.1
KQ459592 KPI96791.1 RSAL01000139 RVE46138.1 AGBW02010564 OWR48298.1 KQ460845 KPJ12002.1 KQ972669 EFA11666.2 GDAI01002166 JAI15437.1 GEZM01071157 JAV65999.1 KK852405 KDR24519.1 ATLV01026175 KE525407 KFB52891.1 GFDL01008875 JAV26170.1 CH477267 EAT45492.1 AAAB01008807 EAA04662.4 APCN01003701 NNAY01000140 OXU30630.1 GGFK01007161 MBW40482.1 CVRI01000010 CRK88773.1 DS231824 EDS27451.1 ADMH02002070 ETN59613.1 AXCN02000985 GGFM01003097 MBW23848.1 GGFM01003092 MBW23843.1 GGFM01003152 MBW23903.1 KA648082 AFP62711.1 GGFJ01003423 MBW52564.1 GGFJ01003309 MBW52450.1 GGFJ01003308 MBW52449.1 AXCM01000491 KB631679 ERL85345.1 CH933806 EDW14738.1 JRES01001623 KNC21417.1 GAKP01011455 JAC47497.1 GBXI01004255 JAD10037.1 KQ434856 KZC08775.1 CH940656 KRF78316.1 CH902617 EDV42747.2 GDHF01016510 JAI35804.1 AF184226 AE014297 AAD55737.1 AAF54976.1 CP012526 ALC46792.1 KQ435720 KOX78634.1 LJIG01022741 KRT78958.1 AJWK01017069 GDHF01007043 JAI45271.1 OUUW01000005 SPP80313.1 GEZM01071155 JAV66001.1 CM000160 KRK03620.1 AAF54977.3 GBYB01004806 GBYB01004807 JAG74573.1 JAG74574.1 GAKP01011450 JAC47502.1 SPP80311.1 CM000070 EDY67874.2 KQ760444 OAD60283.1 CH479179 EDW25000.1 CH916377 EDV90678.1 AJVK01004027 AJVK01004028 AJVK01004029 KRF78313.1 KPU79889.1 GEFH01004461 JAP64120.1 KRK03617.1 ABW08664.1 KZ288222 PBC32038.1 BT003605 AAO39608.1 SPP80314.1 KQ977728 KYN00260.1 GL435766 EFN72743.1 EIM52541.2 QOIP01000008 RLU19977.1 GDRN01038292 JAI67260.1 KQ976396 KYM92889.1 GANP01007215 JAB77253.1 ADTU01011818 KQ982796 KYQ50617.1 KQ981727 KYN37068.1 GEFM01003048 JAP72748.1 KQ981004 KYN10391.1 GAPW01000916 JAC12682.1 GACK01002117 JAA62917.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000007151
UP000053240
+ More
UP000007266 UP000192223 UP000027135 UP000030765 UP000075880 UP000075900 UP000008820 UP000007062 UP000075903 UP000076408 UP000075885 UP000075882 UP000075840 UP000215335 UP000075902 UP000183832 UP000002320 UP000076407 UP000000673 UP000075886 UP000075883 UP000095301 UP000002358 UP000095300 UP000030742 UP000075901 UP000009192 UP000037069 UP000076502 UP000005203 UP000008792 UP000007801 UP000075920 UP000192221 UP000069272 UP000000803 UP000092553 UP000053105 UP000092461 UP000268350 UP000002282 UP000001819 UP000008744 UP000001070 UP000092462 UP000242457 UP000078542 UP000000311 UP000279307 UP000078540 UP000005205 UP000075809 UP000078541 UP000078492
UP000007266 UP000192223 UP000027135 UP000030765 UP000075880 UP000075900 UP000008820 UP000007062 UP000075903 UP000076408 UP000075885 UP000075882 UP000075840 UP000215335 UP000075902 UP000183832 UP000002320 UP000076407 UP000000673 UP000075886 UP000075883 UP000095301 UP000002358 UP000095300 UP000030742 UP000075901 UP000009192 UP000037069 UP000076502 UP000005203 UP000008792 UP000007801 UP000075920 UP000192221 UP000069272 UP000000803 UP000092553 UP000053105 UP000092461 UP000268350 UP000002282 UP000001819 UP000008744 UP000001070 UP000092462 UP000242457 UP000078542 UP000000311 UP000279307 UP000078540 UP000005205 UP000075809 UP000078541 UP000078492
Interpro
Gene 3D
ProteinModelPortal
H9JLC1
A0A2H1X2Z0
A0A2A4K533
A0A194PTQ0
A0A3S2NFE5
A0A212F3I0
+ More
A0A194R2S7 D7EI93 A0A1W4W897 A0A0K8TM45 A0A1Y1KX67 A0A067RXA1 A0A084WRP7 A0A182IS19 A0A182R9U5 A0A1Q3FF44 Q17FY2 A0A1S4F485 Q7QIP3 A0A182UVN1 A0A182Y318 A0A182P131 A0A182L3H7 A0A182I6V5 A0A232FJA9 A0A182TET1 A0A2M4AIC5 A0A1J1HL64 B0W215 A0A182WWL9 W5J893 A0A182QWH2 A0A2M3Z5X9 A0A2M3Z5Q3 A0A2M3Z5V8 T1PGZ9 A0A2M4BHS4 A0A2M4BHB7 A0A2M4BHB0 A0A182LZR7 A0A1I8MZJ6 K7J196 A0A1I8NUC0 U4U5Y7 A0A182S9M1 B4KBR1 A0A0L0BN41 A0A034VZG8 A0A0A1XGE0 A0A154PA54 A0A1I8MZJ8 A0A088AII6 A0A1I8NUB6 A0A0Q9VZN2 B3LWV9 A0A182WHX4 A0A0K8VA22 A0A1W4V588 A0A182FGB9 Q7KSK8 A0A0M4EUK2 A0A0N0BJ46 A0A0T6AVY2 A0A1B0CLB2 A0A0K8W2I9 A0A3B0JHX2 A0A1Y1KZS2 A0A0R1E7F9 Q9V3X1 A0A0C9QVY5 A0A034VZH2 A0A3B0K5Q2 B5DXD8 A0A310SU42 B4G3J2 B4JYB6 A0A1W4USX3 A0A1B0D8B2 A0A0Q9VZN4 A0A0P8ZZ49 A0A131XDE8 A0A0R1E6W6 A8JQZ7 A0A2A3EK13 Q86NW5 A0A3B0JD55 A0A151IGD6 E2A1S0 I5ANK6 A0A3L8DHT2 A0A0P4WRX5 A0A195BW80 V5HSX0 A0A158NCD3 A0A151WS46 A0A195F9E9 A0A131Y2L8 A0A151ITJ9 A0A023EUN6 L7MGG2
A0A194R2S7 D7EI93 A0A1W4W897 A0A0K8TM45 A0A1Y1KX67 A0A067RXA1 A0A084WRP7 A0A182IS19 A0A182R9U5 A0A1Q3FF44 Q17FY2 A0A1S4F485 Q7QIP3 A0A182UVN1 A0A182Y318 A0A182P131 A0A182L3H7 A0A182I6V5 A0A232FJA9 A0A182TET1 A0A2M4AIC5 A0A1J1HL64 B0W215 A0A182WWL9 W5J893 A0A182QWH2 A0A2M3Z5X9 A0A2M3Z5Q3 A0A2M3Z5V8 T1PGZ9 A0A2M4BHS4 A0A2M4BHB7 A0A2M4BHB0 A0A182LZR7 A0A1I8MZJ6 K7J196 A0A1I8NUC0 U4U5Y7 A0A182S9M1 B4KBR1 A0A0L0BN41 A0A034VZG8 A0A0A1XGE0 A0A154PA54 A0A1I8MZJ8 A0A088AII6 A0A1I8NUB6 A0A0Q9VZN2 B3LWV9 A0A182WHX4 A0A0K8VA22 A0A1W4V588 A0A182FGB9 Q7KSK8 A0A0M4EUK2 A0A0N0BJ46 A0A0T6AVY2 A0A1B0CLB2 A0A0K8W2I9 A0A3B0JHX2 A0A1Y1KZS2 A0A0R1E7F9 Q9V3X1 A0A0C9QVY5 A0A034VZH2 A0A3B0K5Q2 B5DXD8 A0A310SU42 B4G3J2 B4JYB6 A0A1W4USX3 A0A1B0D8B2 A0A0Q9VZN4 A0A0P8ZZ49 A0A131XDE8 A0A0R1E6W6 A8JQZ7 A0A2A3EK13 Q86NW5 A0A3B0JD55 A0A151IGD6 E2A1S0 I5ANK6 A0A3L8DHT2 A0A0P4WRX5 A0A195BW80 V5HSX0 A0A158NCD3 A0A151WS46 A0A195F9E9 A0A131Y2L8 A0A151ITJ9 A0A023EUN6 L7MGG2
PDB
4U09
E-value=4.71934e-25,
Score=286
Ontologies
GO
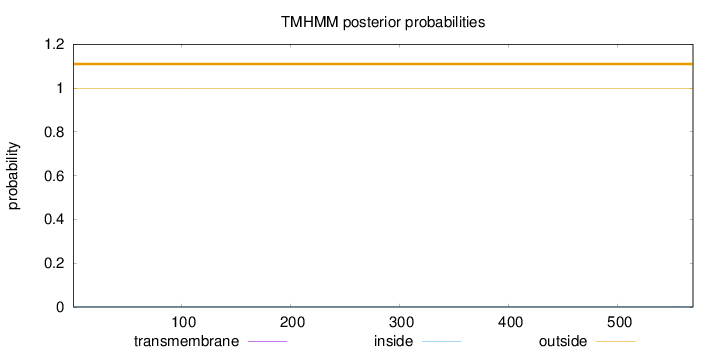
Topology
Length:
569
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0033
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00255
outside
1 - 569
Population Genetic Test Statistics
Pi
24.480201
Theta
21.881606
Tajima's D
0.378808
CLR
0.584632
CSRT
0.48287585620719
Interpretation
Uncertain
