Gene
KWMTBOMO07400 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010324
Annotation
PREDICTED:_uncharacterized_protein_LOC101747119_isoform_X2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.717
Sequence
CDS
ATGAACTCAAACGAAATTATTTATTTTGTAATATTAACATTATGCTCTTCAGTTGCGGGCCAAGTGTGCAGCGACGGGCGGTCGGTGACATTTGTGCGATGGTCCGGAGTGACGTCAGAGCGAGCGCCGCCAGTGTTCGTCTACACGGCTAGCGGCGAAGAAGATTCCCTTGTTGGAGCTTGTTTAGCTCAATGCAGAGAACTGGAGGACTGCGCCGCAGTGGTCGTCGCGTACGGGAAAGGAAATTGTGTCGGGGTAGCTAATACTCCCGAACCCCACTTGAGAACTGATCACGATGTCACGTACTTTAAAAAGATTTGCTTGCATGTTTCCGAAGAATGTTCCACGAAATGGTGGGCGTTAGAGAGTACTCCTGGTTACCATCTACATTCCGAGAGCGCAGTGAAGGTGTTACCAAACTCTACCATCCAAGACTGCTACGATTCAATACTTTCAGCCCGTAGAACTTCATACCGTTCAGGCCAATGGGTGGGACCAGACGGGTCGTTCAACGCGGAGCTGATACAAGCGGCAGATTCAACAGTCGGTAGCTGCCTCGTCAACGCCGAGGATAAGTTCACTGAACCGGAATCTTACAGAGTATCAAACTACTATACATTCTACATCGAGAATCAATGTAAACATGACTATTTAATAAAAATAGATAGATGTTCATACGAAGAATACAACAATCAAACGCTGAGACACGTCGAATTCACACTGAAGAATCGCACCAAAGACGAGTGCAAGAAAGCATGTGAAACGGAACAAAAGTTCATATGTCGAGGATTTAGTTGGATCTCGAAGGAAGAGAAAAGTGGAGTTCGAGGCATTTGCGACTTGCACAGTGAAGATCTTGTTACTACTGGTTCTTGGCTGCTTCGGAAAACGTCAGGTGCAACCTATTATAGGCGGGTCATATGTTTGAATATAAGCGTGGATTGCGACAACAAGGGTTTGAACATAACATACAAGCCGCGCGGAGTTTTTCGGGGTCGTATGTACGTCCCGGGGAAAGGGGAGCAGTGCGGGGCGCGAGGGAAGGGAGGTCCCGTCAAATTGACGTTACCGATGCACGGAGAATGTGACGTTAATTTTGCATACGGAATCTCGAATGGACCTACCGGTGTCATTAATAGAACAATGGCGTACGTAATGGTTATGATCCAGAACAATCCCATAATACAGACGGCAGGAGACAGATGGGTTCGTGTGGGATGTTCGCCTGGACCTCGCACACCCACACATGTAGACGCCACGCTCGCTGTTACTGATAACGGACGGGCCCCAGCGACGAATTCCGGTCGAAAATCGTGGACCGGAGCGGCGAGTGCGGTGGTGGGGGGCAACGCTCCCCTGAGGCTGTACGTGGTACGCGCGGAAACCGTGCAGCCTCCGTCCGCGGCCGTCTCACTCGGGGAACCACTCGAATTGAGGCTTGAGACTACTGATACATCGGAGATTCAACCCTATTATTTAGTGGCCTCATCTCGCCTCGGCGACAGCTCTGTTGTATTGCTGGACAGTAAAGGCTGTCCTACTGGTCAGGTGGATTTCCCGGCATTTACTCGTACCTGGTTGGACGGGACCCAGCGTTTGAGTGTACGGTTCAAAGCGTTCCGTTTTCCTACGTCACACATCGTACGCTTCGCGCTCATGGTGAGATTTTGTGAAGACAAATGCGAAAAGGTAGATTGCATGGAGAATATAGGCAGGATAACTCGTGATGCTAACGTCACGTACTTCAGCGAAGACACGGAGCAGGGTCGCGTCGTGGCGCAGGGAGGGACACGCGTGTGCCACATGGGGGTCGACGACCCTTTGACACTTGAACTAGAGCTGGCCGTAGATTCTAAAGGCATTCAGTCTTCTGACGCACTCGTCAGGGCTGATCATCGGTCAAGCTACCGAATGTTAAGCGTGATTAAAGTTTTCATTTAG
Protein
MNSNEIIYFVILTLCSSVAGQVCSDGRSVTFVRWSGVTSERAPPVFVYTASGEEDSLVGACLAQCRELEDCAAVVVAYGKGNCVGVANTPEPHLRTDHDVTYFKKICLHVSEECSTKWWALESTPGYHLHSESAVKVLPNSTIQDCYDSILSARRTSYRSGQWVGPDGSFNAELIQAADSTVGSCLVNAEDKFTEPESYRVSNYYTFYIENQCKHDYLIKIDRCSYEEYNNQTLRHVEFTLKNRTKDECKKACETEQKFICRGFSWISKEEKSGVRGICDLHSEDLVTTGSWLLRKTSGATYYRRVICLNISVDCDNKGLNITYKPRGVFRGRMYVPGKGEQCGARGKGGPVKLTLPMHGECDVNFAYGISNGPTGVINRTMAYVMVMIQNNPIIQTAGDRWVRVGCSPGPRTPTHVDATLAVTDNGRAPATNSGRKSWTGAASAVVGGNAPLRLYVVRAETVQPPSAAVSLGEPLELRLETTDTSEIQPYYLVASSRLGDSSVVLLDSKGCPTGQVDFPAFTRTWLDGTQRLSVRFKAFRFPTSHIVRFALMVRFCEDKCEKVDCMENIGRITRDANVTYFSEDTEQGRVVAQGGTRVCHMGVDDPLTLELELAVDSKGIQSSDALVRADHRSSYRMLSVIKVFI
Summary
Uniprot
A0A2A4K3D6
A0A2H1W4L3
A0A194R2I6
A0A194PVK1
A0A212F3I1
D6X332
+ More
A0A067RN91 A0A2P8YMN5 A0A2J7PDS5 A0A2J7PDR3 A0A3L8DVC2 A0A0T6BGW1 A0A1B6E728 A0A088AFH9 N6T5M3 A0A232F7T9 A0A1J1HN04 A0A0J7NL95 J9LS79 E1ZYY0 A0A1Y1MXX0 A0A2S2PY34 A0A1W6EWB2 A0A310S483 E2C2K1 A0A224XLL6 K7J4P8 A0A0M4EV08 A0A1Y1N145 B4GP76 B3LV56 A0A195CJ60 A0A023EZN7 A0A1A9Y8K0 A0A3B0K860 B4LZT0 A0A0Q9WGQ1 A0A1A9VE33 A0A1B0F9A8 B4JTG4 A0A026W7N3 F4X6U6 A0A0C9RAM8 B4K4N1 E9J669 A0A0C9R7C8 A0A195E6P4 A0A1A9X2J2 Q29BN3 A0A0N1IU30 B3P7G9 A0A1W4V338 A0A154PGX2 W8BCC6 A0A0L0CRG8 B4PN38 A0A195F1L6 Q9VCR4 A0A158NFW1 A0A195B228 A0A034VP05 A0A0A1WSF6 A0A0K8VH83 A0A0A1XQ87 B4HFA6 A0A151XCA5 A0A087TCV3 A0A1I8NAG5 A0A1S4J279 A0A0K2T1X1 B0W214 A0A1A9ZN42 T1KJV3 Q17FY1 A0A182H5Q1 A0A3R7M4Q0 A0A182H4K1 A0A146LQ91 T1ILR6 A0A226EXA3 A0A1B0CLB1 A0A2P8YR97 A0A2J7R424 A0A2J7R430 A0A0P4WF65 A0A1W6EWC0
A0A067RN91 A0A2P8YMN5 A0A2J7PDS5 A0A2J7PDR3 A0A3L8DVC2 A0A0T6BGW1 A0A1B6E728 A0A088AFH9 N6T5M3 A0A232F7T9 A0A1J1HN04 A0A0J7NL95 J9LS79 E1ZYY0 A0A1Y1MXX0 A0A2S2PY34 A0A1W6EWB2 A0A310S483 E2C2K1 A0A224XLL6 K7J4P8 A0A0M4EV08 A0A1Y1N145 B4GP76 B3LV56 A0A195CJ60 A0A023EZN7 A0A1A9Y8K0 A0A3B0K860 B4LZT0 A0A0Q9WGQ1 A0A1A9VE33 A0A1B0F9A8 B4JTG4 A0A026W7N3 F4X6U6 A0A0C9RAM8 B4K4N1 E9J669 A0A0C9R7C8 A0A195E6P4 A0A1A9X2J2 Q29BN3 A0A0N1IU30 B3P7G9 A0A1W4V338 A0A154PGX2 W8BCC6 A0A0L0CRG8 B4PN38 A0A195F1L6 Q9VCR4 A0A158NFW1 A0A195B228 A0A034VP05 A0A0A1WSF6 A0A0K8VH83 A0A0A1XQ87 B4HFA6 A0A151XCA5 A0A087TCV3 A0A1I8NAG5 A0A1S4J279 A0A0K2T1X1 B0W214 A0A1A9ZN42 T1KJV3 Q17FY1 A0A182H5Q1 A0A3R7M4Q0 A0A182H4K1 A0A146LQ91 T1ILR6 A0A226EXA3 A0A1B0CLB1 A0A2P8YR97 A0A2J7R424 A0A2J7R430 A0A0P4WF65 A0A1W6EWC0
Pubmed
26354079
22118469
18362917
19820115
24845553
29403074
+ More
30249741 23537049 28648823 20798317 28004739 20075255 17994087 25474469 24508170 21719571 21282665 15632085 24495485 26108605 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 25348373 25830018 25315136 17510324 26483478 26823975
30249741 23537049 28648823 20798317 28004739 20075255 17994087 25474469 24508170 21719571 21282665 15632085 24495485 26108605 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 25348373 25830018 25315136 17510324 26483478 26823975
EMBL
NWSH01000173
PCG78771.1
ODYU01006279
SOQ47977.1
KQ460845
KPJ12003.1
+ More
KQ459592 KPI96789.1 AGBW02010564 OWR48299.1 KQ971372 EFA10317.2 KK852405 KDR24518.1 PYGN01000487 PSN45520.1 NEVH01026385 PNF14477.1 PNF14476.1 QOIP01000004 RLU23869.1 LJIG01000392 KRT86548.1 GEDC01018970 GEDC01017225 GEDC01016352 GEDC01014532 GEDC01012726 GEDC01003572 GEDC01001388 JAS18328.1 JAS20073.1 JAS20946.1 JAS22766.1 JAS24572.1 JAS33726.1 JAS35910.1 APGK01043502 APGK01043503 KB741014 ENN75499.1 NNAY01000785 OXU26518.1 CVRI01000010 CRK88772.1 LBMM01003680 KMQ93240.1 ABLF02025740 GL435242 EFN73670.1 GEZM01018909 GEZM01018908 GEZM01018904 GEZM01018902 GEZM01018899 JAV89988.1 GGMS01001252 MBY70455.1 KY563601 ARK20010.1 KQ780658 OAD51932.1 GL452135 EFN77857.1 GFTR01007505 JAW08921.1 AAZX01000464 CP012526 ALC47030.1 GEZM01018905 JAV89976.1 CH479186 EDW38959.1 CH902617 EDV42528.1 KQ977754 KYN00104.1 GBBI01003985 JAC14727.1 OUUW01000005 SPP81201.1 CH940650 EDW68249.2 KRF83753.1 CCAG010018108 CH916373 EDV95054.1 KK107373 EZA51621.1 GL888818 EGI57878.1 GBYB01003881 JAG73648.1 CH933806 EDW15007.2 GL768221 EFZ11694.1 GBYB01003880 GBYB01014035 JAG73647.1 JAG83802.1 KQ979568 KYN20865.1 CM000070 EAL26964.3 KQ435710 KOX79724.1 CH954182 EDV54058.1 KQ434902 KZC11093.1 GAMC01015735 GAMC01015734 GAMC01015733 GAMC01015732 JAB90821.1 JRES01000007 KNC34908.1 CM000160 EDW98093.2 KQ981864 KYN34271.1 AE014297 BT133064 AAF56092.2 AEX93149.1 ADTU01014814 KQ976662 KYM78531.1 GAKP01015680 GAKP01015679 GAKP01015678 JAC43273.1 GBXI01012852 JAD01440.1 GDHF01014073 GDHF01011817 JAI38241.1 JAI40497.1 GBXI01003489 GBXI01001589 JAD10803.1 JAD12703.1 CH480815 EDW43284.1 KQ982314 KYQ57928.1 KK114636 KFM62942.1 HACA01002647 CDW20008.1 DS231824 EDS27450.1 CAEY01000169 CH477267 EAT45493.1 JXUM01112252 JXUM01112253 KQ565576 KXJ70866.1 QCYY01002239 ROT71840.1 JXUM01109795 KQ565354 KXJ71063.1 GDHC01008611 JAQ10018.1 JH430884 LNIX01000001 OXA61704.1 AJWK01017067 AJWK01017068 AJWK01017069 PYGN01000418 PSN46744.1 NEVH01007816 PNF35593.1 PNF35592.1 GDRN01035981 JAI67375.1 KY563603 ARK20012.1
KQ459592 KPI96789.1 AGBW02010564 OWR48299.1 KQ971372 EFA10317.2 KK852405 KDR24518.1 PYGN01000487 PSN45520.1 NEVH01026385 PNF14477.1 PNF14476.1 QOIP01000004 RLU23869.1 LJIG01000392 KRT86548.1 GEDC01018970 GEDC01017225 GEDC01016352 GEDC01014532 GEDC01012726 GEDC01003572 GEDC01001388 JAS18328.1 JAS20073.1 JAS20946.1 JAS22766.1 JAS24572.1 JAS33726.1 JAS35910.1 APGK01043502 APGK01043503 KB741014 ENN75499.1 NNAY01000785 OXU26518.1 CVRI01000010 CRK88772.1 LBMM01003680 KMQ93240.1 ABLF02025740 GL435242 EFN73670.1 GEZM01018909 GEZM01018908 GEZM01018904 GEZM01018902 GEZM01018899 JAV89988.1 GGMS01001252 MBY70455.1 KY563601 ARK20010.1 KQ780658 OAD51932.1 GL452135 EFN77857.1 GFTR01007505 JAW08921.1 AAZX01000464 CP012526 ALC47030.1 GEZM01018905 JAV89976.1 CH479186 EDW38959.1 CH902617 EDV42528.1 KQ977754 KYN00104.1 GBBI01003985 JAC14727.1 OUUW01000005 SPP81201.1 CH940650 EDW68249.2 KRF83753.1 CCAG010018108 CH916373 EDV95054.1 KK107373 EZA51621.1 GL888818 EGI57878.1 GBYB01003881 JAG73648.1 CH933806 EDW15007.2 GL768221 EFZ11694.1 GBYB01003880 GBYB01014035 JAG73647.1 JAG83802.1 KQ979568 KYN20865.1 CM000070 EAL26964.3 KQ435710 KOX79724.1 CH954182 EDV54058.1 KQ434902 KZC11093.1 GAMC01015735 GAMC01015734 GAMC01015733 GAMC01015732 JAB90821.1 JRES01000007 KNC34908.1 CM000160 EDW98093.2 KQ981864 KYN34271.1 AE014297 BT133064 AAF56092.2 AEX93149.1 ADTU01014814 KQ976662 KYM78531.1 GAKP01015680 GAKP01015679 GAKP01015678 JAC43273.1 GBXI01012852 JAD01440.1 GDHF01014073 GDHF01011817 JAI38241.1 JAI40497.1 GBXI01003489 GBXI01001589 JAD10803.1 JAD12703.1 CH480815 EDW43284.1 KQ982314 KYQ57928.1 KK114636 KFM62942.1 HACA01002647 CDW20008.1 DS231824 EDS27450.1 CAEY01000169 CH477267 EAT45493.1 JXUM01112252 JXUM01112253 KQ565576 KXJ70866.1 QCYY01002239 ROT71840.1 JXUM01109795 KQ565354 KXJ71063.1 GDHC01008611 JAQ10018.1 JH430884 LNIX01000001 OXA61704.1 AJWK01017067 AJWK01017068 AJWK01017069 PYGN01000418 PSN46744.1 NEVH01007816 PNF35593.1 PNF35592.1 GDRN01035981 JAI67375.1 KY563603 ARK20012.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000007266
UP000027135
+ More
UP000245037 UP000235965 UP000279307 UP000005203 UP000019118 UP000215335 UP000183832 UP000036403 UP000007819 UP000000311 UP000008237 UP000002358 UP000092553 UP000008744 UP000007801 UP000078542 UP000092443 UP000268350 UP000008792 UP000078200 UP000092444 UP000001070 UP000053097 UP000007755 UP000009192 UP000078492 UP000091820 UP000001819 UP000053105 UP000008711 UP000192221 UP000076502 UP000037069 UP000002282 UP000078541 UP000000803 UP000005205 UP000078540 UP000001292 UP000075809 UP000054359 UP000095301 UP000002320 UP000092445 UP000015104 UP000008820 UP000069940 UP000249989 UP000283509 UP000198287 UP000092461
UP000245037 UP000235965 UP000279307 UP000005203 UP000019118 UP000215335 UP000183832 UP000036403 UP000007819 UP000000311 UP000008237 UP000002358 UP000092553 UP000008744 UP000007801 UP000078542 UP000092443 UP000268350 UP000008792 UP000078200 UP000092444 UP000001070 UP000053097 UP000007755 UP000009192 UP000078492 UP000091820 UP000001819 UP000053105 UP000008711 UP000192221 UP000076502 UP000037069 UP000002282 UP000078541 UP000000803 UP000005205 UP000078540 UP000001292 UP000075809 UP000054359 UP000095301 UP000002320 UP000092445 UP000015104 UP000008820 UP000069940 UP000249989 UP000283509 UP000198287 UP000092461
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4K3D6
A0A2H1W4L3
A0A194R2I6
A0A194PVK1
A0A212F3I1
D6X332
+ More
A0A067RN91 A0A2P8YMN5 A0A2J7PDS5 A0A2J7PDR3 A0A3L8DVC2 A0A0T6BGW1 A0A1B6E728 A0A088AFH9 N6T5M3 A0A232F7T9 A0A1J1HN04 A0A0J7NL95 J9LS79 E1ZYY0 A0A1Y1MXX0 A0A2S2PY34 A0A1W6EWB2 A0A310S483 E2C2K1 A0A224XLL6 K7J4P8 A0A0M4EV08 A0A1Y1N145 B4GP76 B3LV56 A0A195CJ60 A0A023EZN7 A0A1A9Y8K0 A0A3B0K860 B4LZT0 A0A0Q9WGQ1 A0A1A9VE33 A0A1B0F9A8 B4JTG4 A0A026W7N3 F4X6U6 A0A0C9RAM8 B4K4N1 E9J669 A0A0C9R7C8 A0A195E6P4 A0A1A9X2J2 Q29BN3 A0A0N1IU30 B3P7G9 A0A1W4V338 A0A154PGX2 W8BCC6 A0A0L0CRG8 B4PN38 A0A195F1L6 Q9VCR4 A0A158NFW1 A0A195B228 A0A034VP05 A0A0A1WSF6 A0A0K8VH83 A0A0A1XQ87 B4HFA6 A0A151XCA5 A0A087TCV3 A0A1I8NAG5 A0A1S4J279 A0A0K2T1X1 B0W214 A0A1A9ZN42 T1KJV3 Q17FY1 A0A182H5Q1 A0A3R7M4Q0 A0A182H4K1 A0A146LQ91 T1ILR6 A0A226EXA3 A0A1B0CLB1 A0A2P8YR97 A0A2J7R424 A0A2J7R430 A0A0P4WF65 A0A1W6EWC0
A0A067RN91 A0A2P8YMN5 A0A2J7PDS5 A0A2J7PDR3 A0A3L8DVC2 A0A0T6BGW1 A0A1B6E728 A0A088AFH9 N6T5M3 A0A232F7T9 A0A1J1HN04 A0A0J7NL95 J9LS79 E1ZYY0 A0A1Y1MXX0 A0A2S2PY34 A0A1W6EWB2 A0A310S483 E2C2K1 A0A224XLL6 K7J4P8 A0A0M4EV08 A0A1Y1N145 B4GP76 B3LV56 A0A195CJ60 A0A023EZN7 A0A1A9Y8K0 A0A3B0K860 B4LZT0 A0A0Q9WGQ1 A0A1A9VE33 A0A1B0F9A8 B4JTG4 A0A026W7N3 F4X6U6 A0A0C9RAM8 B4K4N1 E9J669 A0A0C9R7C8 A0A195E6P4 A0A1A9X2J2 Q29BN3 A0A0N1IU30 B3P7G9 A0A1W4V338 A0A154PGX2 W8BCC6 A0A0L0CRG8 B4PN38 A0A195F1L6 Q9VCR4 A0A158NFW1 A0A195B228 A0A034VP05 A0A0A1WSF6 A0A0K8VH83 A0A0A1XQ87 B4HFA6 A0A151XCA5 A0A087TCV3 A0A1I8NAG5 A0A1S4J279 A0A0K2T1X1 B0W214 A0A1A9ZN42 T1KJV3 Q17FY1 A0A182H5Q1 A0A3R7M4Q0 A0A182H4K1 A0A146LQ91 T1ILR6 A0A226EXA3 A0A1B0CLB1 A0A2P8YR97 A0A2J7R424 A0A2J7R430 A0A0P4WF65 A0A1W6EWC0
Ontologies
PANTHER
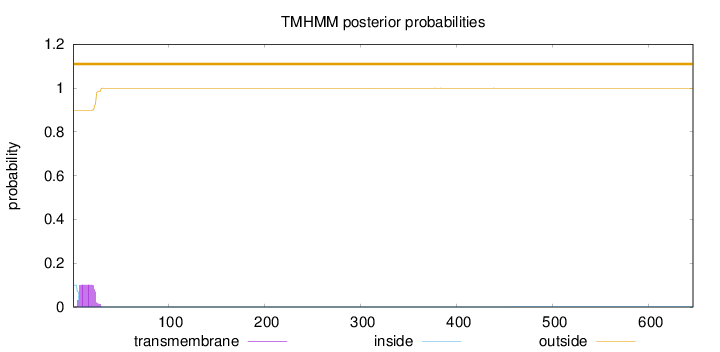
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.933911
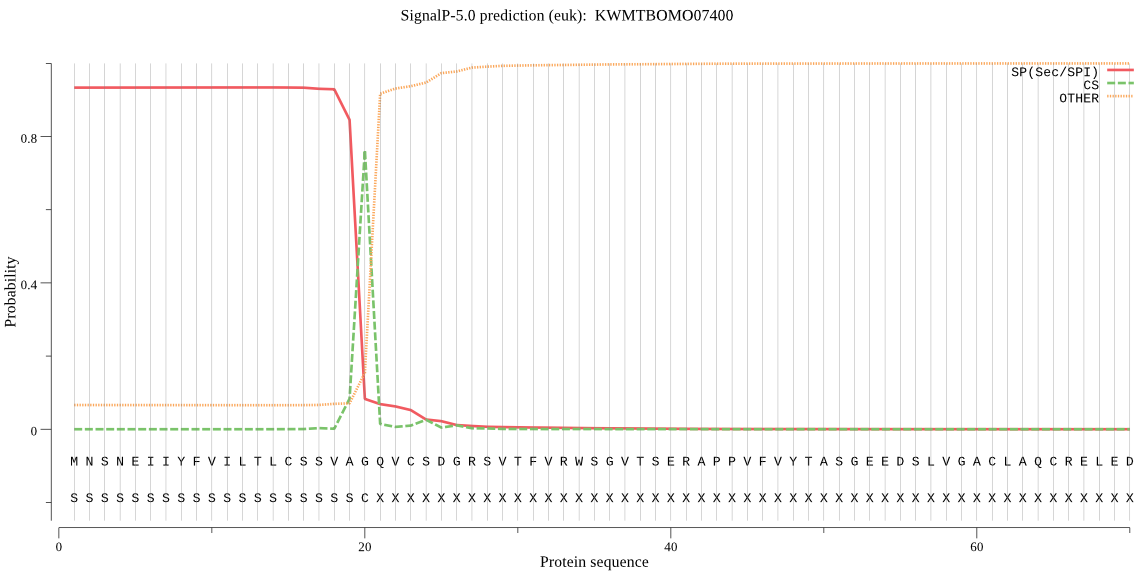
Length:
646
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.91313000000001
Exp number, first 60 AAs:
1.90755
Total prob of N-in:
0.10086
outside
1 - 646
Population Genetic Test Statistics
Pi
15.762408
Theta
17.122889
Tajima's D
-0.939208
CLR
0.578822
CSRT
0.143992800359982
Interpretation
Uncertain
