Gene
KWMTBOMO07394
Pre Gene Modal
BGIBMGA010326
Annotation
PREDICTED:_mitochondrial_basic_amino_acids_transporter-like_isoform_X1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.353 PlasmaMembrane Reliability : 1.208
Sequence
CDS
ATGGCACTAGATTTCGTTGCTGGATGTATTGGAGGTTGTGCGGGAATCGTAGCCGGCCATCCCTTAGACACCCTGAAAGTACACATACAGTCGGGTAGAGGCAGCCCTTTACAATGTGCAAAATCTCTTCTCAGGGGCGGTACGATCAGCAACATATATCGTGGTGTATGGGCTCCACTAGGTGGCATAGCAGCTGTTAACGCAATTGTTTTTGGTGCATACGGAAACACCAGAAGGGCTCTATCGGAACCAGATTCATTAAAGACTCATGCTATGGCGGGAGCAGCAGCGGGCGCTCTCCAGAGTCTAGCTTGTGCACCTGTAGAACTTATTAAAACTCGGCAGCAGCTCGCCAGTGCCGGCGAGGGTATGCCCAATGGAGCCTGGGCTGGAGCGAGACATATAGTAAAGATCGGAGGATTTAAAGCTCTTTTTCGGGGACTTGGAATAACGGTGGCACGTGACAGCCCAGCGTTTGCGATTTATTTTACCTCATACGAGGTTATGACGAGAGGTGATAAATCGGTATCTAGAGTGTTCACAGCGGGCGGGGTGGCGGGGGCACTTTCCTGGGTGATCTTGTATCCGATAGATGTTGTAAAATCACGTTTGCAAGGCGACACTATTGGTAAATACTCTGGTGCTTGGGACTGTTTCGTGAAGTCGATCCGTTCTGACGGATGGCGGTGCATGACCCGAGGCGTGGTTGCTGTGACGCTGCGAGCTTTCATCAGCAACGGCGCGTGCTTCACCGCAGTGGCGTGGACCGAGCAAGCCTGGCAGCATTGCTCATCGGACTCCGCAGTCAAAACGCTGGCACAGGCGGCTGTTATCAGCGACGCAGTGCGGCACCACGAAAGTCCCGACTTACGATTTGACATTTAA
Protein
MALDFVAGCIGGCAGIVAGHPLDTLKVHIQSGRGSPLQCAKSLLRGGTISNIYRGVWAPLGGIAAVNAIVFGAYGNTRRALSEPDSLKTHAMAGAAAGALQSLACAPVELIKTRQQLASAGEGMPNGAWAGARHIVKIGGFKALFRGLGITVARDSPAFAIYFTSYEVMTRGDKSVSRVFTAGGVAGALSWVILYPIDVVKSRLQGDTIGKYSGAWDCFVKSIRSDGWRCMTRGVVAVTLRAFISNGACFTAVAWTEQAWQHCSSDSAVKTLAQAAVISDAVRHHESPDLRFDI
Summary
Similarity
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Uniprot
H9JLC5
I4DL98
A0A194R2T2
A0A0L7LEQ4
A0A2A4K4C2
A0A212F3K4
+ More
A0A3S2NAU4 A0A2H1VW94 A0A026W975 A0A195BV37 A0A195FHT4 A0A195DM93 A0A1Y1KAE3 A0A151WU84 A0A195D4J9 A0A158P2D8 E9IGC1 A0A1B0DF90 K7IWH7 E2AY32 A0A1W4XAY3 F4WUR2 Q16YM1 A0A232F8W8 B0W1I1 A0A1S4FJK5 A0A0A1XB22 A0A1Q3FMM0 A0A1L8DE31 A0A034W649 A0A154PGW5 A0A2J7RCT8 W5JKY0 A0A336M4N5 A0A2M3Z8A5 A0A182F5E1 A0A293LKC8 V5GNX6 A0A2M4BPQ3 A0A2M4BPQ6 A0A1J1IK86 A0A084WBX8 U5EXE6 A0A182I520 A0A182KUL7 A0A182WYQ2 A0A182IJG0 A0A2M4A939 A0A1I8QBG6 Q7PW06 A0A2M4A8C7 A0A1S4H103 A0A182MKX8 A0A2M4A9L7 A0A0K8UT90 A0A182YFC0 A0A182TQE9 A0A182NZV8 A0A1I8MT88 A0A182R9L6 A0A182WH21 T1PAM9 A0A182NEZ9 A0A182VET3 A0A2R5LB26 J3JTB5 A0A0L0CB12 A0A182K5D4 A0A0T6B8L9 A0A1D2NG81 A0A0Q9X6M7 B3MPR3 A0A182Q4N7 A0A023ER56 A0A1W4USX1 W8AX36 A0A1W4UFF3 A0A1W4UFY6 A0A0R1DL78 A0A3R7NCH9 B4NZ98 B3N973 A0A182SK92 E9FV13 A0A3B0K6E4 A0A1B6GS30 A0A3B0K156 A0A1B6M8Q4 B4HWK8 A0A0N0BID8 A0A1B6M3A5 B4Q8T9 A0A0J9R0W8 B4MG48 Q76NQ2 D6X353 A0A0P5QG43 Q8IH44 A0A164V0Y0
A0A3S2NAU4 A0A2H1VW94 A0A026W975 A0A195BV37 A0A195FHT4 A0A195DM93 A0A1Y1KAE3 A0A151WU84 A0A195D4J9 A0A158P2D8 E9IGC1 A0A1B0DF90 K7IWH7 E2AY32 A0A1W4XAY3 F4WUR2 Q16YM1 A0A232F8W8 B0W1I1 A0A1S4FJK5 A0A0A1XB22 A0A1Q3FMM0 A0A1L8DE31 A0A034W649 A0A154PGW5 A0A2J7RCT8 W5JKY0 A0A336M4N5 A0A2M3Z8A5 A0A182F5E1 A0A293LKC8 V5GNX6 A0A2M4BPQ3 A0A2M4BPQ6 A0A1J1IK86 A0A084WBX8 U5EXE6 A0A182I520 A0A182KUL7 A0A182WYQ2 A0A182IJG0 A0A2M4A939 A0A1I8QBG6 Q7PW06 A0A2M4A8C7 A0A1S4H103 A0A182MKX8 A0A2M4A9L7 A0A0K8UT90 A0A182YFC0 A0A182TQE9 A0A182NZV8 A0A1I8MT88 A0A182R9L6 A0A182WH21 T1PAM9 A0A182NEZ9 A0A182VET3 A0A2R5LB26 J3JTB5 A0A0L0CB12 A0A182K5D4 A0A0T6B8L9 A0A1D2NG81 A0A0Q9X6M7 B3MPR3 A0A182Q4N7 A0A023ER56 A0A1W4USX1 W8AX36 A0A1W4UFF3 A0A1W4UFY6 A0A0R1DL78 A0A3R7NCH9 B4NZ98 B3N973 A0A182SK92 E9FV13 A0A3B0K6E4 A0A1B6GS30 A0A3B0K156 A0A1B6M8Q4 B4HWK8 A0A0N0BID8 A0A1B6M3A5 B4Q8T9 A0A0J9R0W8 B4MG48 Q76NQ2 D6X353 A0A0P5QG43 Q8IH44 A0A164V0Y0
Pubmed
19121390
22651552
26354079
26227816
22118469
24508170
+ More
30249741 28004739 21347285 21282665 20075255 20798317 21719571 17510324 28648823 25830018 25348373 20920257 23761445 25765539 24438588 20966253 12364791 25244985 25315136 22516182 26108605 27289101 17994087 24945155 24495485 17550304 21292972 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115
30249741 28004739 21347285 21282665 20075255 20798317 21719571 17510324 28648823 25830018 25348373 20920257 23761445 25765539 24438588 20966253 12364791 25244985 25315136 22516182 26108605 27289101 17994087 24945155 24495485 17550304 21292972 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115
EMBL
BABH01037148
AK402066
KQ459592
BAM18688.1
KPI96785.1
KQ460845
+ More
KPJ12007.1 JTDY01001427 KOB73880.1 NWSH01000173 PCG78758.1 AGBW02010564 OWR48304.1 RSAL01000139 RVE46131.1 ODYU01004825 SOQ45107.1 KK107323 QOIP01000002 EZA52627.1 RLU25521.1 KQ976408 KYM91195.1 KQ981523 KYN40250.1 KQ980734 KYN13947.1 GEZM01087812 JAV58462.1 KQ982753 KYQ51195.1 KQ976885 KYN07354.1 ADTU01007161 ADTU01007162 ADTU01007163 GL762997 EFZ20370.1 AJVK01058619 GL443762 EFN61628.1 GL888375 EGI62059.1 CH477514 EAT39714.1 NNAY01000694 OXU26940.1 DS231822 EDS25779.1 GBXI01007452 GBXI01006327 JAD06840.1 JAD07965.1 GFDL01006185 JAV28860.1 GFDF01009361 JAV04723.1 GAKP01009190 JAC49762.1 KQ434900 KZC11047.1 NEVH01005883 PNF38639.1 ADMH02000797 ETN65032.1 UFQT01000438 SSX24311.1 GGFM01004003 MBW24754.1 GFWV01003589 MAA28319.1 GANP01012388 JAB72080.1 GGFJ01005908 MBW55049.1 GGFJ01005909 MBW55050.1 CVRI01000054 CRL00592.1 ATLV01022517 KE525333 KFB47722.1 GANO01002573 JAB57298.1 APCN01003748 GGFK01003950 MBW37271.1 AAAB01008984 EAA14700.4 GGFK01003733 MBW37054.1 AXCM01001497 GGFK01003987 MBW37308.1 GDHF01022739 JAI29575.1 KA645792 KA649618 AFP60421.1 GGLE01002542 MBY06668.1 BT126471 AEE61435.1 JRES01000648 KNC29608.1 LJIG01009138 KRT83680.1 LJIJ01000049 ODN04271.1 CH964294 KRG00401.1 CH902620 EDV32311.2 AXCN02000838 GAPW01002474 JAC11124.1 GAMC01021169 JAB85386.1 CM000157 KRJ98036.1 QCYY01000677 ROT83498.1 EDW88793.1 KRJ98037.1 CH954177 EDV58508.2 GL732525 EFX88822.1 OUUW01000006 SPP81579.1 GECZ01004540 JAS65229.1 SPP81580.1 GEBQ01007678 JAT32299.1 CH480818 EDW52403.1 KQ435730 KOX77561.1 GEBQ01009599 JAT30378.1 CM000361 EDX04504.1 CM002910 KMY89488.1 KMY89489.1 CH940671 EDW58186.1 AE014134 AAF52912.1 AGB92860.1 KQ971372 EFA10329.2 GDIQ01117601 JAL34125.1 BT001438 AAN71193.1 LRGB01001501 KZS11858.1
KPJ12007.1 JTDY01001427 KOB73880.1 NWSH01000173 PCG78758.1 AGBW02010564 OWR48304.1 RSAL01000139 RVE46131.1 ODYU01004825 SOQ45107.1 KK107323 QOIP01000002 EZA52627.1 RLU25521.1 KQ976408 KYM91195.1 KQ981523 KYN40250.1 KQ980734 KYN13947.1 GEZM01087812 JAV58462.1 KQ982753 KYQ51195.1 KQ976885 KYN07354.1 ADTU01007161 ADTU01007162 ADTU01007163 GL762997 EFZ20370.1 AJVK01058619 GL443762 EFN61628.1 GL888375 EGI62059.1 CH477514 EAT39714.1 NNAY01000694 OXU26940.1 DS231822 EDS25779.1 GBXI01007452 GBXI01006327 JAD06840.1 JAD07965.1 GFDL01006185 JAV28860.1 GFDF01009361 JAV04723.1 GAKP01009190 JAC49762.1 KQ434900 KZC11047.1 NEVH01005883 PNF38639.1 ADMH02000797 ETN65032.1 UFQT01000438 SSX24311.1 GGFM01004003 MBW24754.1 GFWV01003589 MAA28319.1 GANP01012388 JAB72080.1 GGFJ01005908 MBW55049.1 GGFJ01005909 MBW55050.1 CVRI01000054 CRL00592.1 ATLV01022517 KE525333 KFB47722.1 GANO01002573 JAB57298.1 APCN01003748 GGFK01003950 MBW37271.1 AAAB01008984 EAA14700.4 GGFK01003733 MBW37054.1 AXCM01001497 GGFK01003987 MBW37308.1 GDHF01022739 JAI29575.1 KA645792 KA649618 AFP60421.1 GGLE01002542 MBY06668.1 BT126471 AEE61435.1 JRES01000648 KNC29608.1 LJIG01009138 KRT83680.1 LJIJ01000049 ODN04271.1 CH964294 KRG00401.1 CH902620 EDV32311.2 AXCN02000838 GAPW01002474 JAC11124.1 GAMC01021169 JAB85386.1 CM000157 KRJ98036.1 QCYY01000677 ROT83498.1 EDW88793.1 KRJ98037.1 CH954177 EDV58508.2 GL732525 EFX88822.1 OUUW01000006 SPP81579.1 GECZ01004540 JAS65229.1 SPP81580.1 GEBQ01007678 JAT32299.1 CH480818 EDW52403.1 KQ435730 KOX77561.1 GEBQ01009599 JAT30378.1 CM000361 EDX04504.1 CM002910 KMY89488.1 KMY89489.1 CH940671 EDW58186.1 AE014134 AAF52912.1 AGB92860.1 KQ971372 EFA10329.2 GDIQ01117601 JAL34125.1 BT001438 AAN71193.1 LRGB01001501 KZS11858.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000037510
UP000218220
UP000007151
+ More
UP000283053 UP000053097 UP000279307 UP000078540 UP000078541 UP000078492 UP000075809 UP000078542 UP000005205 UP000092462 UP000002358 UP000000311 UP000192223 UP000007755 UP000008820 UP000215335 UP000002320 UP000076502 UP000235965 UP000000673 UP000069272 UP000183832 UP000030765 UP000075840 UP000075882 UP000076407 UP000075880 UP000095300 UP000007062 UP000075883 UP000076408 UP000075902 UP000075885 UP000095301 UP000075900 UP000075920 UP000075884 UP000075903 UP000037069 UP000075881 UP000094527 UP000007798 UP000007801 UP000075886 UP000192221 UP000002282 UP000283509 UP000008711 UP000075901 UP000000305 UP000268350 UP000001292 UP000053105 UP000000304 UP000008792 UP000000803 UP000007266 UP000076858
UP000283053 UP000053097 UP000279307 UP000078540 UP000078541 UP000078492 UP000075809 UP000078542 UP000005205 UP000092462 UP000002358 UP000000311 UP000192223 UP000007755 UP000008820 UP000215335 UP000002320 UP000076502 UP000235965 UP000000673 UP000069272 UP000183832 UP000030765 UP000075840 UP000075882 UP000076407 UP000075880 UP000095300 UP000007062 UP000075883 UP000076408 UP000075902 UP000075885 UP000095301 UP000075900 UP000075920 UP000075884 UP000075903 UP000037069 UP000075881 UP000094527 UP000007798 UP000007801 UP000075886 UP000192221 UP000002282 UP000283509 UP000008711 UP000075901 UP000000305 UP000268350 UP000001292 UP000053105 UP000000304 UP000008792 UP000000803 UP000007266 UP000076858
Pfam
PF00153 Mito_carr
Interpro
SUPFAM
SSF103506
SSF103506
Gene 3D
ProteinModelPortal
H9JLC5
I4DL98
A0A194R2T2
A0A0L7LEQ4
A0A2A4K4C2
A0A212F3K4
+ More
A0A3S2NAU4 A0A2H1VW94 A0A026W975 A0A195BV37 A0A195FHT4 A0A195DM93 A0A1Y1KAE3 A0A151WU84 A0A195D4J9 A0A158P2D8 E9IGC1 A0A1B0DF90 K7IWH7 E2AY32 A0A1W4XAY3 F4WUR2 Q16YM1 A0A232F8W8 B0W1I1 A0A1S4FJK5 A0A0A1XB22 A0A1Q3FMM0 A0A1L8DE31 A0A034W649 A0A154PGW5 A0A2J7RCT8 W5JKY0 A0A336M4N5 A0A2M3Z8A5 A0A182F5E1 A0A293LKC8 V5GNX6 A0A2M4BPQ3 A0A2M4BPQ6 A0A1J1IK86 A0A084WBX8 U5EXE6 A0A182I520 A0A182KUL7 A0A182WYQ2 A0A182IJG0 A0A2M4A939 A0A1I8QBG6 Q7PW06 A0A2M4A8C7 A0A1S4H103 A0A182MKX8 A0A2M4A9L7 A0A0K8UT90 A0A182YFC0 A0A182TQE9 A0A182NZV8 A0A1I8MT88 A0A182R9L6 A0A182WH21 T1PAM9 A0A182NEZ9 A0A182VET3 A0A2R5LB26 J3JTB5 A0A0L0CB12 A0A182K5D4 A0A0T6B8L9 A0A1D2NG81 A0A0Q9X6M7 B3MPR3 A0A182Q4N7 A0A023ER56 A0A1W4USX1 W8AX36 A0A1W4UFF3 A0A1W4UFY6 A0A0R1DL78 A0A3R7NCH9 B4NZ98 B3N973 A0A182SK92 E9FV13 A0A3B0K6E4 A0A1B6GS30 A0A3B0K156 A0A1B6M8Q4 B4HWK8 A0A0N0BID8 A0A1B6M3A5 B4Q8T9 A0A0J9R0W8 B4MG48 Q76NQ2 D6X353 A0A0P5QG43 Q8IH44 A0A164V0Y0
A0A3S2NAU4 A0A2H1VW94 A0A026W975 A0A195BV37 A0A195FHT4 A0A195DM93 A0A1Y1KAE3 A0A151WU84 A0A195D4J9 A0A158P2D8 E9IGC1 A0A1B0DF90 K7IWH7 E2AY32 A0A1W4XAY3 F4WUR2 Q16YM1 A0A232F8W8 B0W1I1 A0A1S4FJK5 A0A0A1XB22 A0A1Q3FMM0 A0A1L8DE31 A0A034W649 A0A154PGW5 A0A2J7RCT8 W5JKY0 A0A336M4N5 A0A2M3Z8A5 A0A182F5E1 A0A293LKC8 V5GNX6 A0A2M4BPQ3 A0A2M4BPQ6 A0A1J1IK86 A0A084WBX8 U5EXE6 A0A182I520 A0A182KUL7 A0A182WYQ2 A0A182IJG0 A0A2M4A939 A0A1I8QBG6 Q7PW06 A0A2M4A8C7 A0A1S4H103 A0A182MKX8 A0A2M4A9L7 A0A0K8UT90 A0A182YFC0 A0A182TQE9 A0A182NZV8 A0A1I8MT88 A0A182R9L6 A0A182WH21 T1PAM9 A0A182NEZ9 A0A182VET3 A0A2R5LB26 J3JTB5 A0A0L0CB12 A0A182K5D4 A0A0T6B8L9 A0A1D2NG81 A0A0Q9X6M7 B3MPR3 A0A182Q4N7 A0A023ER56 A0A1W4USX1 W8AX36 A0A1W4UFF3 A0A1W4UFY6 A0A0R1DL78 A0A3R7NCH9 B4NZ98 B3N973 A0A182SK92 E9FV13 A0A3B0K6E4 A0A1B6GS30 A0A3B0K156 A0A1B6M8Q4 B4HWK8 A0A0N0BID8 A0A1B6M3A5 B4Q8T9 A0A0J9R0W8 B4MG48 Q76NQ2 D6X353 A0A0P5QG43 Q8IH44 A0A164V0Y0
PDB
2LCK
E-value=4.83975e-07,
Score=127
Ontologies
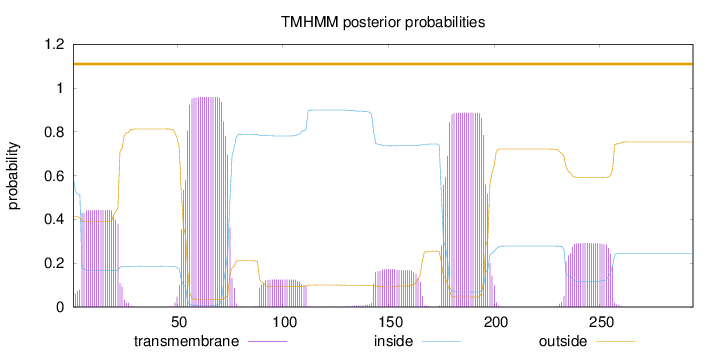
Topology
Length:
294
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
60.4880500000001
Exp number, first 60 AAs:
15.47655
Total prob of N-in:
0.58760
POSSIBLE N-term signal
sequence
outside
1 - 294
Population Genetic Test Statistics
Pi
18.039134
Theta
14.476929
Tajima's D
0.796611
CLR
1.922365
CSRT
0.614469276536173
Interpretation
Uncertain
