Gene
KWMTBOMO07392
Pre Gene Modal
BGIBMGA010339
Annotation
PREDICTED:_kelch-like_protein_10_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.676
Sequence
CDS
ATGATGATAACGGTGCACTGGCAACAGCAGACGGCGGGTCGGGAAACCGGTCGGTGCATGTCGGTGGCAGCGATGCAGGCGCTCCATCGACTGCGCGAGTGTCGTCTGCTATGCGACGCTATTGTGCGCGCTGACGATGGCGCTTCCTTCCCCGTGCACCGTGCTATACTCTCAGCCTGCAGCCCATACTTCCTGGCACTGTTCACAACAACTCTTCATTCTCGGGAGCAAAGCGACGTATTGATATCCGGAGTGCGTTCTGAGATATTACTGCTGTTGATTGAGTACGCCTACCTCCGCCGTATCGATGTCACAGATACCAATGTACATGAGCTCCTAATGACAGCCGATTATCTCGCTTTCCTTGGAGTGCTTCAGCTTTGTTGCGATCATCTCAGAAGTTCATTGAACCCAAAAAACTGTCTTGGAATTATGACTTTTGCAAGGCGTGTTTTCTGTTACAAACTCGAAGCGGATGCACGAAGGTATTTATTGCGTTACTTTGTAACAGTCGCAACACAGAGCGACGAATTCCTTCACTTGCCACTGGACGAACTTAATTCTATTATATTGGAGGACGAATTAAACGTTAAAAGTGAGGAAGCAGTTTGGGAATCCGTTTTGCGTTGGGTCAATTACGATCCAGACGGTCGATGGCAGCACACGGTAAAATTAATGGGCAGCATTCGACTGGGATTGCTCGACACGCAATTCTTCCTAGAGAATGTTAAAGATCATCCATACGTTGCCGGGAATGAAGGATCTAGGCCAGTTATAATAGAGACGCTGAAGTTCTTATACGATTTGGAAATGATTGCTCAGAGAGATGGAGAAGTTGCCACACCAGAAATAGCCCGACCAAGAGTGCCGCATGAGGTCTTGTTTGCCATTGGAGGATGGAGTGGTGGGTCTCCGACTGCTTTCATCGAAACCTACGATACAAGAGCTGATCGCTGGATCAAAGTCGAAGAAGTGGATCCGGCTGGTCCACGAGCCTACCACGGAACAGCAGTCTTAGGATCTTGCATATACGTTATCGGCGGTTTTGACGGCATGGACTACTTCAACTCGTGTCGCTGTTTTGATGCCGTCACAAAAATATGGCGCGAGGTTGCACCCATGAACGCAAGGCGTTGTTACGTGTCTGTTGCTGTCGTTGGTGAGACAATCTATGCAATGGGCGGTTACGATGGCCACCACAGACAGAACACGGCCGAAAAGTTTAATCATCGAACGAATCAATGGTCGCTGGTGGCACCGATGAATGCACAACGTTCCGATGCAAGCGCCGCCTCACTTGACAACAAGATATACATAACCGGTGGCTTCAATGGTCAGGAATGCATGAACAGCGTCGAAATGTACGATCCAGAAGCGAACCAATGGACGAATTTGACCCCGATGAGGTCGCGCCGCTCCGGCGTGTCCTGCATCGCTTACCACAACAAGGTCTACGTCATTGGAGGCTTCAACGGTATCTCGAGGATGTGCAGCGGCGAGGTGTACGACCCGACGACCAACACGTGGGCGCCCGTGCCGGACATGTACAATCCCCGCAGCAACTTTGCCATCGAGGTCATTGACGACATGATCTTTGCGATCGGCGGCTTCAATGGAGTCACCACTATATACCACGTCGAGTGCTACGACGAGAAGACGAACGAGTGGTGA
Protein
MMITVHWQQQTAGRETGRCMSVAAMQALHRLRECRLLCDAIVRADDGASFPVHRAILSACSPYFLALFTTTLHSREQSDVLISGVRSEILLLLIEYAYLRRIDVTDTNVHELLMTADYLAFLGVLQLCCDHLRSSLNPKNCLGIMTFARRVFCYKLEADARRYLLRYFVTVATQSDEFLHLPLDELNSIILEDELNVKSEEAVWESVLRWVNYDPDGRWQHTVKLMGSIRLGLLDTQFFLENVKDHPYVAGNEGSRPVIIETLKFLYDLEMIAQRDGEVATPEIARPRVPHEVLFAIGGWSGGSPTAFIETYDTRADRWIKVEEVDPAGPRAYHGTAVLGSCIYVIGGFDGMDYFNSCRCFDAVTKIWREVAPMNARRCYVSVAVVGETIYAMGGYDGHHRQNTAEKFNHRTNQWSLVAPMNAQRSDASAASLDNKIYITGGFNGQECMNSVEMYDPEANQWTNLTPMRSRRSGVSCIAYHNKVYVIGGFNGISRMCSGEVYDPTTNTWAPVPDMYNPRSNFAIEVIDDMIFAIGGFNGVTTIYHVECYDEKTNEW
Summary
Uniprot
H9JLD8
A0A2H1VWD3
A0A2A4K3C9
A0A3S2PAR6
A0A212F3J7
A0A0L7LEQ9
+ More
A0A2A4K3X6 A0A2A4K526 A0A2A3EI13 A0A088A9H5 A0A0L7RH03 A0A195DM84 A0A151WTN2 A0A195D368 A0A195FJR8 E2AY31 A0A195BTD2 A0A158P2D9 A0A026W9J9 A0A224X953 A0A154PTU6 T1HSE5 A0A1W4X0E7 E0VNX1 D6X354 A0A310SHK4 U4U558 A0A1Y1NHD7 N6TUA4 A0A0A9YPI4 A0A0A9YMX4 A0A0K8THF7 A0A0M9A5C5 A0A0C9QNW1 A0A1B6FBT9 A0A1B6GHM5 A0A0C9PMI8 A0A1B6L0N1 A0A1D2NDK7 K7IWH8 A0A3L8DYD6 A0A336K738 A0A067QSH7 A0A1B0FR17 A0A1A9ZFC0 A0A226ELQ3 A0A2P8YSC4 J9K0R4 V4B2M3 A0A034V7S1 V4BZ38 W8AMQ1 W8BKG4 A0A1I8NBE4 A0A0K8WCD1 A0A0A1WY65 A0A2J7Q7T1 A0A1A9VF41 A0A2J7PMB4 B4KFF4 A0A2J7R9U5 A0A034VB60 W8BAE0 B4LQP6 B4JQ27 A0A2C9KFA9 A0A2C9KF84 K1QZM1 A0A0T6B8M1 A0A210QRR8 I5AMP5 A0A0Q9WQ92 A0A210QKN9 A0A0P8Y7U2 A0A2C9JDU9 A0A1S4FFK7 Q6GKZ1 Q173I2 A0A3B0JWV1 A0A3S1AGY3 A0A067RFF4 A0A1W4UG57 A0A0Q9X4N1 A0A0Q9X5I3 A0A0Q9WJR0 A0A0Q9WIJ7 A0A1I8PCH9 A0A1Q3FT70 B0W9S6 A0A1J1IRB7 A0A0K8W6Q9 A0A210QDN8 A0A067QVB8 A0A182SQN7 A0A1B6D3P4 A0A0P9BUX1 A0A067QQP0 K1R9Z0
A0A2A4K3X6 A0A2A4K526 A0A2A3EI13 A0A088A9H5 A0A0L7RH03 A0A195DM84 A0A151WTN2 A0A195D368 A0A195FJR8 E2AY31 A0A195BTD2 A0A158P2D9 A0A026W9J9 A0A224X953 A0A154PTU6 T1HSE5 A0A1W4X0E7 E0VNX1 D6X354 A0A310SHK4 U4U558 A0A1Y1NHD7 N6TUA4 A0A0A9YPI4 A0A0A9YMX4 A0A0K8THF7 A0A0M9A5C5 A0A0C9QNW1 A0A1B6FBT9 A0A1B6GHM5 A0A0C9PMI8 A0A1B6L0N1 A0A1D2NDK7 K7IWH8 A0A3L8DYD6 A0A336K738 A0A067QSH7 A0A1B0FR17 A0A1A9ZFC0 A0A226ELQ3 A0A2P8YSC4 J9K0R4 V4B2M3 A0A034V7S1 V4BZ38 W8AMQ1 W8BKG4 A0A1I8NBE4 A0A0K8WCD1 A0A0A1WY65 A0A2J7Q7T1 A0A1A9VF41 A0A2J7PMB4 B4KFF4 A0A2J7R9U5 A0A034VB60 W8BAE0 B4LQP6 B4JQ27 A0A2C9KFA9 A0A2C9KF84 K1QZM1 A0A0T6B8M1 A0A210QRR8 I5AMP5 A0A0Q9WQ92 A0A210QKN9 A0A0P8Y7U2 A0A2C9JDU9 A0A1S4FFK7 Q6GKZ1 Q173I2 A0A3B0JWV1 A0A3S1AGY3 A0A067RFF4 A0A1W4UG57 A0A0Q9X4N1 A0A0Q9X5I3 A0A0Q9WJR0 A0A0Q9WIJ7 A0A1I8PCH9 A0A1Q3FT70 B0W9S6 A0A1J1IRB7 A0A0K8W6Q9 A0A210QDN8 A0A067QVB8 A0A182SQN7 A0A1B6D3P4 A0A0P9BUX1 A0A067QQP0 K1R9Z0
Pubmed
19121390
22118469
26227816
20798317
21347285
24508170
+ More
20566863 18362917 19820115 23537049 28004739 25401762 27289101 20075255 30249741 24845553 29403074 23254933 25348373 24495485 25315136 25830018 17994087 15562597 22992520 28812685 15632085 18057021 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
20566863 18362917 19820115 23537049 28004739 25401762 27289101 20075255 30249741 24845553 29403074 23254933 25348373 24495485 25315136 25830018 17994087 15562597 22992520 28812685 15632085 18057021 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01037148
ODYU01004825
SOQ45108.1
NWSH01000173
PCG78761.1
RSAL01000139
+ More
RVE46130.1 AGBW02010564 OWR48305.1 JTDY01001427 KOB73885.1 PCG78759.1 PCG78760.1 KZ288254 PBC30839.1 KQ414594 KOC70140.1 KQ980734 KYN13946.1 KQ982753 KYQ51196.1 KQ976885 KYN07355.1 KQ981523 KYN40249.1 GL443762 EFN61627.1 KQ976408 KYM91196.1 ADTU01007164 ADTU01007165 KK107323 EZA52628.1 GFTR01007560 JAW08866.1 KQ435093 KZC14620.1 ACPB03004813 DS235354 EEB15077.1 KQ971372 EFA10330.1 KQ759885 OAD62157.1 KB632133 ERL89019.1 GEZM01002346 JAV97312.1 APGK01018556 KB740081 ENN81608.1 GBHO01010073 GBRD01001242 JAG33531.1 JAG64579.1 GBHO01010075 JAG33529.1 GBRD01001241 JAG64580.1 KQ435730 KOX77560.1 GBYB01002312 JAG72079.1 GECZ01022092 JAS47677.1 GECZ01007843 JAS61926.1 GBYB01002313 JAG72080.1 GEBQ01022893 JAT17084.1 LJIJ01000077 ODN03344.1 QOIP01000002 RLU25302.1 UFQS01000048 UFQT01000048 SSW98573.1 SSX18959.1 KK853004 KDR12596.1 CCAG010000954 LNIX01000003 OXA57496.1 PYGN01000390 PSN47135.1 ABLF02030839 ABLF02030841 ABLF02043432 ABLF02058034 KB200701 ESP00682.1 GAKP01019616 JAC39336.1 KB201813 ESO94394.1 GAMC01016510 JAB90045.1 GAMC01016511 JAB90044.1 GDHF01003592 JAI48722.1 GBXI01010520 JAD03772.1 NEVH01017333 PNF24644.1 NEVH01024210 PNF17473.1 CH933807 EDW12054.1 NEVH01006580 PNF37608.1 GAKP01019615 JAC39337.1 GAMC01016509 JAB90046.1 CH940649 EDW63430.1 CH916372 EDV99007.1 JH818378 EKC42522.1 LJIG01009138 KRT83679.1 NEDP02002269 OWF51388.1 CH379061 EIM52230.1 CH963857 KRF98403.1 NEDP02003184 OWF49315.1 CH902628 KPU75324.1 KPU75325.1 BT014923 BT022090 CP007081 AAT47774.1 AAY33506.2 EAA46324.2 EAA46325.2 CH477424 EAT41190.1 OUUW01000010 SPP85553.1 RQTK01000005 RUS91794.1 KK852541 KDR21753.1 KRG03060.1 KRG03062.1 KRG03061.1 KRF81115.1 KRF81116.1 KRF81117.1 KRF81119.1 GFDL01004251 JAV30794.1 DS231865 EDS40395.1 CVRI01000058 CRL02759.1 GDHF01005729 JAI46585.1 NEDP02004067 OWF46852.1 KK852899 KDR14136.1 GEDC01028608 GEDC01016987 JAS08690.1 JAS20311.1 KPU75326.1 KK853081 KDR11680.1 EKC42523.1
RVE46130.1 AGBW02010564 OWR48305.1 JTDY01001427 KOB73885.1 PCG78759.1 PCG78760.1 KZ288254 PBC30839.1 KQ414594 KOC70140.1 KQ980734 KYN13946.1 KQ982753 KYQ51196.1 KQ976885 KYN07355.1 KQ981523 KYN40249.1 GL443762 EFN61627.1 KQ976408 KYM91196.1 ADTU01007164 ADTU01007165 KK107323 EZA52628.1 GFTR01007560 JAW08866.1 KQ435093 KZC14620.1 ACPB03004813 DS235354 EEB15077.1 KQ971372 EFA10330.1 KQ759885 OAD62157.1 KB632133 ERL89019.1 GEZM01002346 JAV97312.1 APGK01018556 KB740081 ENN81608.1 GBHO01010073 GBRD01001242 JAG33531.1 JAG64579.1 GBHO01010075 JAG33529.1 GBRD01001241 JAG64580.1 KQ435730 KOX77560.1 GBYB01002312 JAG72079.1 GECZ01022092 JAS47677.1 GECZ01007843 JAS61926.1 GBYB01002313 JAG72080.1 GEBQ01022893 JAT17084.1 LJIJ01000077 ODN03344.1 QOIP01000002 RLU25302.1 UFQS01000048 UFQT01000048 SSW98573.1 SSX18959.1 KK853004 KDR12596.1 CCAG010000954 LNIX01000003 OXA57496.1 PYGN01000390 PSN47135.1 ABLF02030839 ABLF02030841 ABLF02043432 ABLF02058034 KB200701 ESP00682.1 GAKP01019616 JAC39336.1 KB201813 ESO94394.1 GAMC01016510 JAB90045.1 GAMC01016511 JAB90044.1 GDHF01003592 JAI48722.1 GBXI01010520 JAD03772.1 NEVH01017333 PNF24644.1 NEVH01024210 PNF17473.1 CH933807 EDW12054.1 NEVH01006580 PNF37608.1 GAKP01019615 JAC39337.1 GAMC01016509 JAB90046.1 CH940649 EDW63430.1 CH916372 EDV99007.1 JH818378 EKC42522.1 LJIG01009138 KRT83679.1 NEDP02002269 OWF51388.1 CH379061 EIM52230.1 CH963857 KRF98403.1 NEDP02003184 OWF49315.1 CH902628 KPU75324.1 KPU75325.1 BT014923 BT022090 CP007081 AAT47774.1 AAY33506.2 EAA46324.2 EAA46325.2 CH477424 EAT41190.1 OUUW01000010 SPP85553.1 RQTK01000005 RUS91794.1 KK852541 KDR21753.1 KRG03060.1 KRG03062.1 KRG03061.1 KRF81115.1 KRF81116.1 KRF81117.1 KRF81119.1 GFDL01004251 JAV30794.1 DS231865 EDS40395.1 CVRI01000058 CRL02759.1 GDHF01005729 JAI46585.1 NEDP02004067 OWF46852.1 KK852899 KDR14136.1 GEDC01028608 GEDC01016987 JAS08690.1 JAS20311.1 KPU75326.1 KK853081 KDR11680.1 EKC42523.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000037510
UP000242457
+ More
UP000005203 UP000053825 UP000078492 UP000075809 UP000078542 UP000078541 UP000000311 UP000078540 UP000005205 UP000053097 UP000076502 UP000015103 UP000192223 UP000009046 UP000007266 UP000030742 UP000019118 UP000053105 UP000094527 UP000002358 UP000279307 UP000027135 UP000092444 UP000092445 UP000198287 UP000245037 UP000007819 UP000030746 UP000095301 UP000235965 UP000078200 UP000009192 UP000008792 UP000001070 UP000076420 UP000005408 UP000242188 UP000001819 UP000007798 UP000007801 UP000000803 UP000008820 UP000268350 UP000271974 UP000192221 UP000095300 UP000002320 UP000183832 UP000075901
UP000005203 UP000053825 UP000078492 UP000075809 UP000078542 UP000078541 UP000000311 UP000078540 UP000005205 UP000053097 UP000076502 UP000015103 UP000192223 UP000009046 UP000007266 UP000030742 UP000019118 UP000053105 UP000094527 UP000002358 UP000279307 UP000027135 UP000092444 UP000092445 UP000198287 UP000245037 UP000007819 UP000030746 UP000095301 UP000235965 UP000078200 UP000009192 UP000008792 UP000001070 UP000076420 UP000005408 UP000242188 UP000001819 UP000007798 UP000007801 UP000000803 UP000008820 UP000268350 UP000271974 UP000192221 UP000095300 UP000002320 UP000183832 UP000075901
Interpro
Gene 3D
ProteinModelPortal
H9JLD8
A0A2H1VWD3
A0A2A4K3C9
A0A3S2PAR6
A0A212F3J7
A0A0L7LEQ9
+ More
A0A2A4K3X6 A0A2A4K526 A0A2A3EI13 A0A088A9H5 A0A0L7RH03 A0A195DM84 A0A151WTN2 A0A195D368 A0A195FJR8 E2AY31 A0A195BTD2 A0A158P2D9 A0A026W9J9 A0A224X953 A0A154PTU6 T1HSE5 A0A1W4X0E7 E0VNX1 D6X354 A0A310SHK4 U4U558 A0A1Y1NHD7 N6TUA4 A0A0A9YPI4 A0A0A9YMX4 A0A0K8THF7 A0A0M9A5C5 A0A0C9QNW1 A0A1B6FBT9 A0A1B6GHM5 A0A0C9PMI8 A0A1B6L0N1 A0A1D2NDK7 K7IWH8 A0A3L8DYD6 A0A336K738 A0A067QSH7 A0A1B0FR17 A0A1A9ZFC0 A0A226ELQ3 A0A2P8YSC4 J9K0R4 V4B2M3 A0A034V7S1 V4BZ38 W8AMQ1 W8BKG4 A0A1I8NBE4 A0A0K8WCD1 A0A0A1WY65 A0A2J7Q7T1 A0A1A9VF41 A0A2J7PMB4 B4KFF4 A0A2J7R9U5 A0A034VB60 W8BAE0 B4LQP6 B4JQ27 A0A2C9KFA9 A0A2C9KF84 K1QZM1 A0A0T6B8M1 A0A210QRR8 I5AMP5 A0A0Q9WQ92 A0A210QKN9 A0A0P8Y7U2 A0A2C9JDU9 A0A1S4FFK7 Q6GKZ1 Q173I2 A0A3B0JWV1 A0A3S1AGY3 A0A067RFF4 A0A1W4UG57 A0A0Q9X4N1 A0A0Q9X5I3 A0A0Q9WJR0 A0A0Q9WIJ7 A0A1I8PCH9 A0A1Q3FT70 B0W9S6 A0A1J1IRB7 A0A0K8W6Q9 A0A210QDN8 A0A067QVB8 A0A182SQN7 A0A1B6D3P4 A0A0P9BUX1 A0A067QQP0 K1R9Z0
A0A2A4K3X6 A0A2A4K526 A0A2A3EI13 A0A088A9H5 A0A0L7RH03 A0A195DM84 A0A151WTN2 A0A195D368 A0A195FJR8 E2AY31 A0A195BTD2 A0A158P2D9 A0A026W9J9 A0A224X953 A0A154PTU6 T1HSE5 A0A1W4X0E7 E0VNX1 D6X354 A0A310SHK4 U4U558 A0A1Y1NHD7 N6TUA4 A0A0A9YPI4 A0A0A9YMX4 A0A0K8THF7 A0A0M9A5C5 A0A0C9QNW1 A0A1B6FBT9 A0A1B6GHM5 A0A0C9PMI8 A0A1B6L0N1 A0A1D2NDK7 K7IWH8 A0A3L8DYD6 A0A336K738 A0A067QSH7 A0A1B0FR17 A0A1A9ZFC0 A0A226ELQ3 A0A2P8YSC4 J9K0R4 V4B2M3 A0A034V7S1 V4BZ38 W8AMQ1 W8BKG4 A0A1I8NBE4 A0A0K8WCD1 A0A0A1WY65 A0A2J7Q7T1 A0A1A9VF41 A0A2J7PMB4 B4KFF4 A0A2J7R9U5 A0A034VB60 W8BAE0 B4LQP6 B4JQ27 A0A2C9KFA9 A0A2C9KF84 K1QZM1 A0A0T6B8M1 A0A210QRR8 I5AMP5 A0A0Q9WQ92 A0A210QKN9 A0A0P8Y7U2 A0A2C9JDU9 A0A1S4FFK7 Q6GKZ1 Q173I2 A0A3B0JWV1 A0A3S1AGY3 A0A067RFF4 A0A1W4UG57 A0A0Q9X4N1 A0A0Q9X5I3 A0A0Q9WJR0 A0A0Q9WIJ7 A0A1I8PCH9 A0A1Q3FT70 B0W9S6 A0A1J1IRB7 A0A0K8W6Q9 A0A210QDN8 A0A067QVB8 A0A182SQN7 A0A1B6D3P4 A0A0P9BUX1 A0A067QQP0 K1R9Z0
PDB
5YQ4
E-value=4.15474e-52,
Score=519
Ontologies
GO
Topology
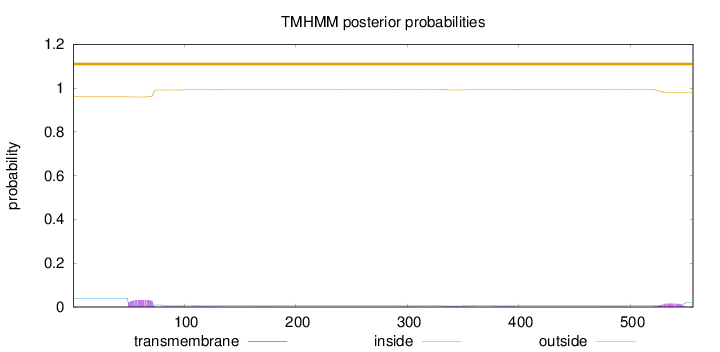
Length:
556
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.21486
Exp number, first 60 AAs:
0.31406
Total prob of N-in:
0.04024
outside
1 - 556
Population Genetic Test Statistics
Pi
17.714941
Theta
15.670676
Tajima's D
0.145728
CLR
0.445696
CSRT
0.416579171041448
Interpretation
Uncertain
