Pre Gene Modal
BGIBMGA010337
Annotation
PREDICTED:_RNA_polymerase_II_transcriptional_coactivator_[Papilio_machaon]
Full name
RNA polymerase II transcriptional coactivator
Alternative Name
p11
Transcription factor
Location in the cell
Nuclear Reliability : 3.501
Sequence
CDS
ATGCCTAAACAAAAGAGAGAAGAAAAACCTAAGAACAAGAAAAACGATAGTTCTAGTAGTGACAGTGATGACGGCCCTGAAGATAGGAACCCTCCTGCTGAAAAAAAGGCAAAAATGGCAGACAGGACTAATGATAAAGAGCCAACATGGGTGTTACAGGGCAAGAAATTACTGAAAGTCCGAGAATTCAAAGGAAAAGTATATGTAGACATAAGAGAATTTTATGAAAAGAATGGTGAACTTTTACCAGGTAAAAAAGGGATAAGTCTCACTCCAGAACAATGGAGAAAATTGTTATCAGTAGGTGAAGAGGTTAATGAGACTGTTAGTTCCATGTGTTAA
Protein
MPKQKREEKPKNKKNDSSSSDSDDGPEDRNPPAEKKAKMADRTNDKEPTWVLQGKKLLKVREFKGKVYVDIREFYEKNGELLPGKKGISLTPEQWRKLLSVGEEVNETVSSMC
Summary
Description
General coactivator that functions cooperatively with TAFs and mediates functional interactions between upstream activators and the general transcriptional machinery. Binds single-stranded DNA (By similarity). Binds specifically to the NssBF element, a short nucleotide sequence of the 1731 retrotransposon, to repress promoter activity.
Similarity
Belongs to the transcriptional coactivator PC4 family.
Belongs to the DEAD box helicase family.
Belongs to the DEAD box helicase family.
Keywords
Activator
Complete proteome
DNA-binding
Nucleus
Reference proteome
Repressor
Transcription
Transcription regulation
Feature
chain RNA polymerase II transcriptional coactivator
Uniprot
A0A2H1V6G4
A0A194R2T6
A0A2A4K4C6
A0A3S2LWX8
A0A194PTP0
I4DL99
+ More
A0A212F3J1 A0A1E1WSB6 A9CTQ5 S4P875 A0A0L7L4S2 A0A067QUX2 A0A0L0C5L1 A0A2P8XGM8 R4WJP6 A0A2S2QF21 A0A1B0D2B4 B4NWF3 A0A1W4UV16 A8D8C5 B3N720 U5EXJ9 A0A2J7R3R7 A0A0M5IW01 A0A1Q3F3I1 A0A023ECV9 A0A2R7WFZ5 B0WYK9 A0A1B0CJZ2 W8AVU4 A0A2M3ZDM0 A0A1A9UY98 A0A1I8PWB1 A0A1A9Z9F3 A0A023EET7 J9JYF1 A0A1A9Y509 A0A1B0AWX8 Q16XW9 A0A2M3ZKB7 A0A0K8UB16 A0A164S6X0 A0A1B2AJ64 Q9VLR5 A0A0P5FD41 A0A2M4C4A8 B4JB34 N6STT5 A0A0P4XBI1 A0A2M4ATI4 A0A023ECU3 A0A2M4C494 B4HYG4 B4Q6B6 W5JGS1 A0A1I8M2C4 B4MVX6 A0A1B0FHM8 A0A2S2NVE8 A0A182FT97 A0A2H8TVV2 B3MPH5 A0A1W4WUX3 A0A226EW24 A0A1D2MXD6 A0A1L8EBS8 B4LUB6 A0A1L8EC48 A0A1S3DHG8 B4KGW9 A0A1B6IMP3 A0A1B6KUG9 A0A0P4VT06 T1H8G2 A0A293M4Q4 D6WW15 A0A1A9VZR5 A0A034WI66 A0A1Y1LXT4 A0A195B259 A0A1B6F020 A0A195E6D3 A0A0P4WKV7 G3MRI6 A0A182Q9D1 A0A023GDN4 Q29ML7 B4G919
A0A212F3J1 A0A1E1WSB6 A9CTQ5 S4P875 A0A0L7L4S2 A0A067QUX2 A0A0L0C5L1 A0A2P8XGM8 R4WJP6 A0A2S2QF21 A0A1B0D2B4 B4NWF3 A0A1W4UV16 A8D8C5 B3N720 U5EXJ9 A0A2J7R3R7 A0A0M5IW01 A0A1Q3F3I1 A0A023ECV9 A0A2R7WFZ5 B0WYK9 A0A1B0CJZ2 W8AVU4 A0A2M3ZDM0 A0A1A9UY98 A0A1I8PWB1 A0A1A9Z9F3 A0A023EET7 J9JYF1 A0A1A9Y509 A0A1B0AWX8 Q16XW9 A0A2M3ZKB7 A0A0K8UB16 A0A164S6X0 A0A1B2AJ64 Q9VLR5 A0A0P5FD41 A0A2M4C4A8 B4JB34 N6STT5 A0A0P4XBI1 A0A2M4ATI4 A0A023ECU3 A0A2M4C494 B4HYG4 B4Q6B6 W5JGS1 A0A1I8M2C4 B4MVX6 A0A1B0FHM8 A0A2S2NVE8 A0A182FT97 A0A2H8TVV2 B3MPH5 A0A1W4WUX3 A0A226EW24 A0A1D2MXD6 A0A1L8EBS8 B4LUB6 A0A1L8EC48 A0A1S3DHG8 B4KGW9 A0A1B6IMP3 A0A1B6KUG9 A0A0P4VT06 T1H8G2 A0A293M4Q4 D6WW15 A0A1A9VZR5 A0A034WI66 A0A1Y1LXT4 A0A195B259 A0A1B6F020 A0A195E6D3 A0A0P4WKV7 G3MRI6 A0A182Q9D1 A0A023GDN4 Q29ML7 B4G919
Pubmed
EMBL
ODYU01000923
SOQ36421.1
KQ460845
KPJ12012.1
NWSH01000173
PCG78764.1
+ More
RSAL01000139 RVE46127.1 KQ459592 KPI96781.1 AK402067 BAM18689.1 AGBW02010564 OWR48308.1 GDQN01001115 JAT89939.1 AB371909 BAF95583.1 GAIX01006066 JAA86494.1 JTDY01003002 KOB70331.1 KK852921 KDR13791.1 JRES01000864 KNC27683.1 PYGN01002208 PSN31161.1 AK417800 BAN21015.1 GGMS01006917 MBY76120.1 AJVK01022719 CM000157 EDW88470.1 EU142272 ABV60408.1 CH954177 EDV59316.1 GANO01000917 JAB58954.1 NEVH01007818 PNF35476.1 CP012523 ALC38206.1 GFDL01012935 JAV22110.1 GAPW01006391 JAC07207.1 KK854773 PTY18587.1 DS232191 EDS37086.1 AJWK01015541 GAMC01021654 JAB84901.1 GGFM01005854 MBW26605.1 GAPW01006389 GAPW01006388 JAC07209.1 ABLF02015899 JXJN01004961 CH477530 EAT39475.1 EJY57774.1 GGFM01008149 MBW28900.1 GDHF01028551 JAI23763.1 LRGB01002076 KZS09316.1 KX531387 ANY27197.1 U18971 AE014134 BT023261 GDIQ01256790 JAJ94934.1 GGFJ01010993 MBW60134.1 CH916368 EDW03926.1 APGK01056555 KB741277 ENN71074.1 GDIP01243430 JAI79971.1 GGFK01010794 MBW44115.1 GAPW01006390 JAC07208.1 GGFJ01010992 MBW60133.1 CH480818 EDW52094.1 CM000361 CM002910 EDX04192.1 KMY88978.1 ADMH02001578 ETN61999.1 CH963857 EDW75846.1 CCAG010020379 GGMR01008403 MBY21022.1 GFXV01006235 MBW18040.1 CH902620 EDV31271.2 LNIX01000001 OXA61719.1 LJIJ01000418 ODM97703.1 GFDG01002599 JAV16200.1 CH940649 EDW64102.2 GFDG01002607 JAV16192.1 CH933807 EDW11169.1 GECU01019527 JAS88179.1 GEBQ01025083 JAT14894.1 GDKW01000265 JAI56330.1 ACPB03015821 GFWV01010342 MAA35071.1 KQ971361 EFA08648.1 GAKP01003716 JAC55236.1 GEZM01045908 GEZM01045907 JAV77628.1 KQ976662 KYM78377.1 GECZ01026221 JAS43548.1 KQ979568 KYN20733.1 GDRN01013365 JAI67861.1 JO844487 AEO36104.1 AXCN02001663 GBBM01003132 JAC32286.1 CH379060 EAL33676.2 CH479180 EDW28849.1
RSAL01000139 RVE46127.1 KQ459592 KPI96781.1 AK402067 BAM18689.1 AGBW02010564 OWR48308.1 GDQN01001115 JAT89939.1 AB371909 BAF95583.1 GAIX01006066 JAA86494.1 JTDY01003002 KOB70331.1 KK852921 KDR13791.1 JRES01000864 KNC27683.1 PYGN01002208 PSN31161.1 AK417800 BAN21015.1 GGMS01006917 MBY76120.1 AJVK01022719 CM000157 EDW88470.1 EU142272 ABV60408.1 CH954177 EDV59316.1 GANO01000917 JAB58954.1 NEVH01007818 PNF35476.1 CP012523 ALC38206.1 GFDL01012935 JAV22110.1 GAPW01006391 JAC07207.1 KK854773 PTY18587.1 DS232191 EDS37086.1 AJWK01015541 GAMC01021654 JAB84901.1 GGFM01005854 MBW26605.1 GAPW01006389 GAPW01006388 JAC07209.1 ABLF02015899 JXJN01004961 CH477530 EAT39475.1 EJY57774.1 GGFM01008149 MBW28900.1 GDHF01028551 JAI23763.1 LRGB01002076 KZS09316.1 KX531387 ANY27197.1 U18971 AE014134 BT023261 GDIQ01256790 JAJ94934.1 GGFJ01010993 MBW60134.1 CH916368 EDW03926.1 APGK01056555 KB741277 ENN71074.1 GDIP01243430 JAI79971.1 GGFK01010794 MBW44115.1 GAPW01006390 JAC07208.1 GGFJ01010992 MBW60133.1 CH480818 EDW52094.1 CM000361 CM002910 EDX04192.1 KMY88978.1 ADMH02001578 ETN61999.1 CH963857 EDW75846.1 CCAG010020379 GGMR01008403 MBY21022.1 GFXV01006235 MBW18040.1 CH902620 EDV31271.2 LNIX01000001 OXA61719.1 LJIJ01000418 ODM97703.1 GFDG01002599 JAV16200.1 CH940649 EDW64102.2 GFDG01002607 JAV16192.1 CH933807 EDW11169.1 GECU01019527 JAS88179.1 GEBQ01025083 JAT14894.1 GDKW01000265 JAI56330.1 ACPB03015821 GFWV01010342 MAA35071.1 KQ971361 EFA08648.1 GAKP01003716 JAC55236.1 GEZM01045908 GEZM01045907 JAV77628.1 KQ976662 KYM78377.1 GECZ01026221 JAS43548.1 KQ979568 KYN20733.1 GDRN01013365 JAI67861.1 JO844487 AEO36104.1 AXCN02001663 GBBM01003132 JAC32286.1 CH379060 EAL33676.2 CH479180 EDW28849.1
Proteomes
UP000053240
UP000218220
UP000283053
UP000053268
UP000007151
UP000037510
+ More
UP000027135 UP000037069 UP000245037 UP000092462 UP000002282 UP000192221 UP000008711 UP000235965 UP000092553 UP000002320 UP000092461 UP000078200 UP000095300 UP000092445 UP000007819 UP000092443 UP000092460 UP000008820 UP000076858 UP000000803 UP000001070 UP000019118 UP000001292 UP000000304 UP000000673 UP000095301 UP000007798 UP000092444 UP000069272 UP000007801 UP000192223 UP000198287 UP000094527 UP000008792 UP000079169 UP000009192 UP000015103 UP000007266 UP000091820 UP000078540 UP000078492 UP000075886 UP000001819 UP000008744
UP000027135 UP000037069 UP000245037 UP000092462 UP000002282 UP000192221 UP000008711 UP000235965 UP000092553 UP000002320 UP000092461 UP000078200 UP000095300 UP000092445 UP000007819 UP000092443 UP000092460 UP000008820 UP000076858 UP000000803 UP000001070 UP000019118 UP000001292 UP000000304 UP000000673 UP000095301 UP000007798 UP000092444 UP000069272 UP000007801 UP000192223 UP000198287 UP000094527 UP000008792 UP000079169 UP000009192 UP000015103 UP000007266 UP000091820 UP000078540 UP000078492 UP000075886 UP000001819 UP000008744
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2H1V6G4
A0A194R2T6
A0A2A4K4C6
A0A3S2LWX8
A0A194PTP0
I4DL99
+ More
A0A212F3J1 A0A1E1WSB6 A9CTQ5 S4P875 A0A0L7L4S2 A0A067QUX2 A0A0L0C5L1 A0A2P8XGM8 R4WJP6 A0A2S2QF21 A0A1B0D2B4 B4NWF3 A0A1W4UV16 A8D8C5 B3N720 U5EXJ9 A0A2J7R3R7 A0A0M5IW01 A0A1Q3F3I1 A0A023ECV9 A0A2R7WFZ5 B0WYK9 A0A1B0CJZ2 W8AVU4 A0A2M3ZDM0 A0A1A9UY98 A0A1I8PWB1 A0A1A9Z9F3 A0A023EET7 J9JYF1 A0A1A9Y509 A0A1B0AWX8 Q16XW9 A0A2M3ZKB7 A0A0K8UB16 A0A164S6X0 A0A1B2AJ64 Q9VLR5 A0A0P5FD41 A0A2M4C4A8 B4JB34 N6STT5 A0A0P4XBI1 A0A2M4ATI4 A0A023ECU3 A0A2M4C494 B4HYG4 B4Q6B6 W5JGS1 A0A1I8M2C4 B4MVX6 A0A1B0FHM8 A0A2S2NVE8 A0A182FT97 A0A2H8TVV2 B3MPH5 A0A1W4WUX3 A0A226EW24 A0A1D2MXD6 A0A1L8EBS8 B4LUB6 A0A1L8EC48 A0A1S3DHG8 B4KGW9 A0A1B6IMP3 A0A1B6KUG9 A0A0P4VT06 T1H8G2 A0A293M4Q4 D6WW15 A0A1A9VZR5 A0A034WI66 A0A1Y1LXT4 A0A195B259 A0A1B6F020 A0A195E6D3 A0A0P4WKV7 G3MRI6 A0A182Q9D1 A0A023GDN4 Q29ML7 B4G919
A0A212F3J1 A0A1E1WSB6 A9CTQ5 S4P875 A0A0L7L4S2 A0A067QUX2 A0A0L0C5L1 A0A2P8XGM8 R4WJP6 A0A2S2QF21 A0A1B0D2B4 B4NWF3 A0A1W4UV16 A8D8C5 B3N720 U5EXJ9 A0A2J7R3R7 A0A0M5IW01 A0A1Q3F3I1 A0A023ECV9 A0A2R7WFZ5 B0WYK9 A0A1B0CJZ2 W8AVU4 A0A2M3ZDM0 A0A1A9UY98 A0A1I8PWB1 A0A1A9Z9F3 A0A023EET7 J9JYF1 A0A1A9Y509 A0A1B0AWX8 Q16XW9 A0A2M3ZKB7 A0A0K8UB16 A0A164S6X0 A0A1B2AJ64 Q9VLR5 A0A0P5FD41 A0A2M4C4A8 B4JB34 N6STT5 A0A0P4XBI1 A0A2M4ATI4 A0A023ECU3 A0A2M4C494 B4HYG4 B4Q6B6 W5JGS1 A0A1I8M2C4 B4MVX6 A0A1B0FHM8 A0A2S2NVE8 A0A182FT97 A0A2H8TVV2 B3MPH5 A0A1W4WUX3 A0A226EW24 A0A1D2MXD6 A0A1L8EBS8 B4LUB6 A0A1L8EC48 A0A1S3DHG8 B4KGW9 A0A1B6IMP3 A0A1B6KUG9 A0A0P4VT06 T1H8G2 A0A293M4Q4 D6WW15 A0A1A9VZR5 A0A034WI66 A0A1Y1LXT4 A0A195B259 A0A1B6F020 A0A195E6D3 A0A0P4WKV7 G3MRI6 A0A182Q9D1 A0A023GDN4 Q29ML7 B4G919
PDB
4USG
E-value=3.16643e-08,
Score=132
Ontologies
GO
Topology
Subcellular location
Nucleus
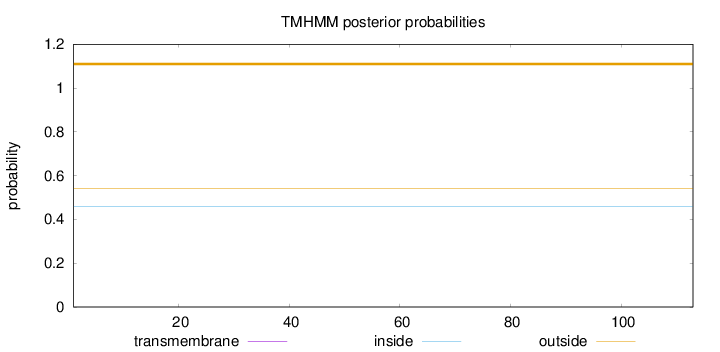
Length:
113
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.45975
outside
1 - 113
Population Genetic Test Statistics
Pi
6.845674
Theta
13.111557
Tajima's D
-1.174509
CLR
5.124936
CSRT
0.104494775261237
Interpretation
Uncertain
