Gene
KWMTBOMO07383 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010330
Annotation
PREDICTED:_multidrug_resistance-associated_protein_1_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.492
Sequence
CDS
ATGTCTTACAACTCCACGCTTGACAGCTTTTGCGGGACACCGTTTTGGAACAGTACTGCCACCTGGTACACGGATAATCCTGAACTAACACCATGTTTCCAGCAGACTGTCCTCATTTGGACACCCTGTCTCTATCTCTGGGTGTTTGCATTCCTTGACTTATATTACATATTCAATAGTAAGGAACGAAATATACCATGGAACATACTGAACATCACGAAATTGCTAGTAACATGTCTTTTAATAGTTTTGAAATTTGTGGACTTGGGTGTAGCAGTCCATTTATCTTCAAACGGAGAAAAGGAAGTCTACAATGCGAATTATTACAGTCCTGTAATAAAAATATTAACATTCGGCTTATCAGCAACATTGCTATTCTATAACAGAAAGTATGGTATGAGAGCATCTGGAGTGCTCTTTTTCTTCTGGTTACTGCTCGTCGTAGCTGGAATACCCCAGCTAAGATCCGAAATTATCGATCACAAGAATTTAGATGACGACGAAAACGTCAAATACAATTTTATTAGTTACATGGTCTACTATCCTTTGATAGTTCTCATGTTCATACTTAATTGTTTTGCCGATTTGCCACCTAAGGACACGCCTTATAAGTATCAAAAGAATCAATGTCCGGAGAATGCAGCGGGATTCCCGAGCCGCCTCACGTTTAGCTGGTTCGATCCGCTCGCCCTGACTGGATTCCGCAGGAGTCTTGTTGAGAACGATCTGTGGGCCCTGAACCCACCGGACTCTTCGAAAGAATGCGTTCCGAAATTCGATAAATTTTGGGAAAGATCTCTGAAGAAACGCGAACTATCGAACGGTACTAAGGCGACATACCGAAAAACTTCAGCCAGTGTCAACTTCAAGCCAGAAAATGAAAAGAAGCCAGCCTCGATATTACCAGCACTGTGTCTCGCATTTGGAGGACAGTTTCTTTTTGGTTCTATCTTGAAGTTAATCAACGATATTCTCATGTTTATATCTCCACAGCTCCTCAAATTATTAATAAGTTTCGTTAAGAACGATGAACCGGACTGGAAAGGTTACGCATACGCCGTCGCGCTGCTGCTGTGTGCCATTACTCAAACAATGCTATTGGCGCACTACTTCACTAGGATGTATCTAGTGGGTATGAGGATCAGAACAGCGTTGACGAGCGCCATCTACAGAAAATCCCTACGTTTGTCGAACTCGGCTAGAAAGGAGTCAACCGTTGGAGAAATTGTCAATTTGATGTCTGTCGATGCTCAGAGATTCGTTGAGTTAACGGCATATTTGAACATGATATGGTCGGCGCCTCTGCAGATCGCCCTCGCCCTATATTTCTTGTGGGCTATCTTAGGTCCGTCAGTTCTCGCTGGTTTGGCGGTAATGATCATTCTTATACCGGTGAACGGTCTAATAGCGAACAGGGTCAAGACATTGCAAATAAAACAGATGAAGTACAAGGATGAGAGAGTTAAACTTATGAACGAAGTGCTCAATGGAATTAAGGTTCTGAAAATGTACGCTTGGGAGCCTAGCTTCGAAGATCAGATCCTGCAAATAAGGAACAAGGAAATGCACGTACTGAAACAAACGGCGTATCTAAATTCTGCGACCTCTTTCATTTGGTCATGTGCTCCGTTTTTGGTTTCACTGATGTCGTTCGGATGCTTTGTATTGGTGAACGATAAGGAAACTCTGGACTCTGAGAAGGCTTTCGTTGCATTATCACTTTTCAACATACTGCGTTTTCCACTGTCAATGCTGCCTAACGTTATATCGAATGTAGTACAAACGTCGGTAGGCATAAAGAGATTGAACAAATTCATGAACTGCGATGAGCTCGACATTAGTTCCGTCGATCACGACAAGAAAGAGCCCAGTCCAATTGTTATCGAAAACGGGAATTTCACGTGGGGCGAAAAAGATGCCGATCCTGTATTGAAGAACATTAATTTGAATGTACCGCGTGGCTCTTTGGTGGCCATAGTCGGAGCTGTAGGTTCCGGGAAGAGTTCATTGCTCGCTGCAATGCTTGGCGAGATGAACAAGATATCAGGCAGAGTGAATACACACGGCAGCATAGCGTACGTCCCGCAACAGGCCTGGATCCAAAATGCCACGCTACAAGATAATATACTCTTTGGGAAGCCTCTGCAGCAGCAGTCTTATAATAACGTTATTAATGTGTGCGCCCTCAAACCCGACTTTGACGTGTTACCTGGAGGAGATCAGACCGAAATCGGAGAGAAAGGTATAAACTTATCGGGCGGTCAGAAGCAGCGCGTGTCGCTGGCGCGCGCCGTGTACCACGAGGCGGACAACTACCTGCTGGACGACCCGCTGAGCGCCGTGGACTCGCACGTCGGCAAGCACATCTTCGACAAGGTCATCGGGCCCGCCGGCCTGCTCAAGGACAGGACGCGCGTCTGGGTCACGCACAACGTGTCCTACCTCGCGCAGACCGACCTGGTCGTCGTGCTGCGCGACGGACAGGTCTCCGAGGCCGGCTCCTACCAGCACCTGCTCGAGAAGAAGGGCGCCTTCGCTGACTTCCTGCTTCATCATCTGAGCGACATCGAGAAAACTTCGCCTGACGAGTTAGATGTTCTCAAGCAAGACCTGGAAACAAAACTCGGTACAGAATTCCAGAACAAACTGCAAAGGGCTCGCTCGCTCTCCGAAAGTACTTCAGAGTCGGAACAAACTCCCGCAGGTGATAGAGCTGGCAGCGTGAAACAAAAGACTCCTGACGCCCTCACTCAAAGCAATCTCAAAGAAAAGAACAAACTGATTGAAGCCGAGAAAGCGGAAACAGGAAGTGTGAAATGGAGTATATATAAGCACTACTTGATGAGCGTCGGTGTTTTCGCGTCCGTCGTGACGATTCTGATGAATCTGATCCTGCAAGTGTTCCAAGTCGGCTCCAACTACTGGCTGGCGGAGTGGTCCAGTGATTCGAAAATCATTGTTAACGGAACAGTGGACAGAGCAAAAAGGGACATGTACCTCGGTGTGTATGGAGCGCTCGGAGCAGGACAGGTGCTGTCGGTGTCGGTGTCGTCGCTGGCGCTATACCTGGGCACGCTGGCGGCGGCGCGCGCCCTCCACGCGACTCTGCTGGCCGGCGTGCTGCGGGCGCCGTCCATCGGCTTCTTCGACTGCACGCCCGTCGGCCGCGTGCTCAACCGCTTCAGCAAGGACGTTGACGTGCTCGACAACACGCTGCCCATGACGCTGCGGGGCTGGACGTCCTGCTTCTTCTCGGTACTGGGAACGCTGTTCGTGATAAGCGTGTCGACGCCCTTATTCTTAGTGATCGTGGTCCCGGTGGGTCTGATCTACTACGTGATCCAGAGGTTCTACGTGGCGACGTCCCGGCAACTCAAACGGCTGGAGTCGATATCGCGATCGCCCATCTATTCGCATTTCGGCGAGAGTATCACCGGGGCGTCCACAATACGAGCCTACGGTGTAACTGATAGATTTATTGAAGAAAGCGAAAGTCGAGTGGATCACAATCAGTCGTGCTATTACCCGAGCTGCATCGCGAATCGGTGGCTGGCCATCCGTCTGGAAATGATTGGAAACTTCATTATATTCAGCGCGGCTGTGTTCGCGGTGCTCGGTAGAAACTCTATCTATCCGGGTATTGTAGGACTCTCGATCAGCTATGCGCTACAGATCACTCAAACACTCAATTGGTTGGTCAGAATGACCTCTGAAGTTGAAACGAATATTGTCGCCGTCGAAAGGATAAAAGAGTACGCGGAAACCGAACAGGAGGCCGCGTGGAACTTGGAGAAAGGGCCCGGCGCAACGTGGCCCGAGACGGGAGCGCTACAGCTGGAGCAGCTGACGCTGGAGTACCGCCCCGGCGAGCCGGCGCTGCGTGACGTCACGTGCACGGTGGCGCCGCGCGACAAGCTCGGCATCGTGGGACGCACCGGCGCCGGCAAGTCTACTCTCACGCTCGGGCTCTTCAGGATCGTGGAGGCAGCCGCCGGACGCATCCTGATCGACGGCGTCGACATCGCGACCCTCGGGCTACACCAGCTGCGGTCGCGCATCACCATCATCCCGCAGGAGCCGATCCTGTTCTCGGGCACGCTGCGCTCCAACCTGGACCCGTTCGAGGCGTACAGCGACGAGGAGATCTGGCGCGCGCTGGAACACGCCCACCTCCGTGCCTTCGTGCAGGGGCTGCCGGCCGGGCTGCGGCACGAGGTGGCGGAGGGCGGCGAGAACCTGTCGGTGGGACAGCGCCAGCTCGTGTGCCTGGCGCGCGCGCTGCTACGCAAGACGCCGCTGCTGGTGCTGGACGAGGCCACCGCGGCCGTCGACCTCGAGACTGACGACCTCATCCAGAAGACGATCCGCTCGGAGTTCGCGTCGTGCACGGTGCTCACTATCGCGCACCGCCTCAACACCATCATGGACTCCACCAAAGTGATGGTGCTCGACCGCGGGCAGCTCGTAGAATATGCGGCCCCTCAACAACTACTTAACGATAAAAACTCCATTTTCTACTCCATGGCTAAAGATGCCGGAATAGTTAACTAA
Protein
MSYNSTLDSFCGTPFWNSTATWYTDNPELTPCFQQTVLIWTPCLYLWVFAFLDLYYIFNSKERNIPWNILNITKLLVTCLLIVLKFVDLGVAVHLSSNGEKEVYNANYYSPVIKILTFGLSATLLFYNRKYGMRASGVLFFFWLLLVVAGIPQLRSEIIDHKNLDDDENVKYNFISYMVYYPLIVLMFILNCFADLPPKDTPYKYQKNQCPENAAGFPSRLTFSWFDPLALTGFRRSLVENDLWALNPPDSSKECVPKFDKFWERSLKKRELSNGTKATYRKTSASVNFKPENEKKPASILPALCLAFGGQFLFGSILKLINDILMFISPQLLKLLISFVKNDEPDWKGYAYAVALLLCAITQTMLLAHYFTRMYLVGMRIRTALTSAIYRKSLRLSNSARKESTVGEIVNLMSVDAQRFVELTAYLNMIWSAPLQIALALYFLWAILGPSVLAGLAVMIILIPVNGLIANRVKTLQIKQMKYKDERVKLMNEVLNGIKVLKMYAWEPSFEDQILQIRNKEMHVLKQTAYLNSATSFIWSCAPFLVSLMSFGCFVLVNDKETLDSEKAFVALSLFNILRFPLSMLPNVISNVVQTSVGIKRLNKFMNCDELDISSVDHDKKEPSPIVIENGNFTWGEKDADPVLKNINLNVPRGSLVAIVGAVGSGKSSLLAAMLGEMNKISGRVNTHGSIAYVPQQAWIQNATLQDNILFGKPLQQQSYNNVINVCALKPDFDVLPGGDQTEIGEKGINLSGGQKQRVSLARAVYHEADNYLLDDPLSAVDSHVGKHIFDKVIGPAGLLKDRTRVWVTHNVSYLAQTDLVVVLRDGQVSEAGSYQHLLEKKGAFADFLLHHLSDIEKTSPDELDVLKQDLETKLGTEFQNKLQRARSLSESTSESEQTPAGDRAGSVKQKTPDALTQSNLKEKNKLIEAEKAETGSVKWSIYKHYLMSVGVFASVVTILMNLILQVFQVGSNYWLAEWSSDSKIIVNGTVDRAKRDMYLGVYGALGAGQVLSVSVSSLALYLGTLAAARALHATLLAGVLRAPSIGFFDCTPVGRVLNRFSKDVDVLDNTLPMTLRGWTSCFFSVLGTLFVISVSTPLFLVIVVPVGLIYYVIQRFYVATSRQLKRLESISRSPIYSHFGESITGASTIRAYGVTDRFIEESESRVDHNQSCYYPSCIANRWLAIRLEMIGNFIIFSAAVFAVLGRNSIYPGIVGLSISYALQITQTLNWLVRMTSEVETNIVAVERIKEYAETEQEAAWNLEKGPGATWPETGALQLEQLTLEYRPGEPALRDVTCTVAPRDKLGIVGRTGAGKSTLTLGLFRIVEAAAGRILIDGVDIATLGLHQLRSRITIIPQEPILFSGTLRSNLDPFEAYSDEEIWRALEHAHLRAFVQGLPAGLRHEVAEGGENLSVGQRQLVCLARALLRKTPLLVLDEATAAVDLETDDLIQKTIRSEFASCTVLTIAHRLNTIMDSTKVMVLDRGQLVEYAAPQQLLNDKNSIFYSMAKDAGIVN
Summary
Uniprot
A0A2H4KAX1
A0A0F6PP97
A0A220A455
D2Y5V6
A0A0F6PQJ9
A0A3S2NW63
+ More
K7J1C7 A0A026W727 A0A139WC08 A0A1W4XSV3 A0A1Y1N5M9 A0A1Y1N973 A0A1W4XTX1 A0A1L8DUP5 A0A1W4XTS4 A0A182H2B8 A0A1W4XIR5 A0A1B6EGY9 A0A1B6E1F3 A0A1Q3FWR2 A0A1S4F8S7 A0A0M8ZP27 A0A1J1IJS3 A0A161ANL7 A0A023F4D8 A0A069DZT4 A0A2A3EBU1 A0A2S2QJQ0 A0A154PCK7 A0A146M0N2 X1WW89 A0A125RM75 A0A1I8PGD2 A0A1I8M696 A0A1I8PGK1 A0A1I8M6A3 A0A1I8PGC3 A0A1I8PGJ1 A0A1I8M6A1 A0A1I8M699 A0A0Q9XGP2 A0A1I8M694 A0A1I8M691 A0A1I8M687 A0A1I8M6A2 A0A1I8PGK7 A0A1I8PGL6 T1PG89 A0A034V5U5 A0A1I8PGB6 T1PED2 A0A1I8PGJ8 A0A1I8PGC8 A0A1I8M698 A0A1I8PGB1 A0A1I8PGB4 A0A1I8PGJ2 A0A1I8M6A6 A0A034V2K4 A0A1I8M6B5 A0A1I8M692 A0A336KSQ1 A0A034V6X7 A0A0Q9X3T7 A0A0Q9XDD3 A0A0Q9XEN4 A0A0Q9WL68 A0A1B0F930 A0A1I8PGK2 A0A1I8PGB9 A0A0Q9X475 N6UIL0 A0A0Q9WLL0 E2B8D1 A0A0Q9X3Q7 A0A0Q9XGA4 A0A336MD69 A0A1I8PGL1 A0A1I8M6A8 A0A1I8M6A4 A0A1I8M6B3 A0A0Q9WXE7 A0A0Q9WYS7 A0A1I8M6B0 A0A0Q9WWH8 A0A0Q9WLR3 A0A1I8PGC2 A0A0Q9WMZ8 A0A336KWU5 A0A1I8PGK6 A0A1I8M6B1 A0A1I8PGJ6 A0A0Q9X4D4 A0A1I8PGK3 B4KIM7 A0A1I8M6A9 A0A1Q3FWN9 A0A336MEP5 A0A0R3P0Y9
K7J1C7 A0A026W727 A0A139WC08 A0A1W4XSV3 A0A1Y1N5M9 A0A1Y1N973 A0A1W4XTX1 A0A1L8DUP5 A0A1W4XTS4 A0A182H2B8 A0A1W4XIR5 A0A1B6EGY9 A0A1B6E1F3 A0A1Q3FWR2 A0A1S4F8S7 A0A0M8ZP27 A0A1J1IJS3 A0A161ANL7 A0A023F4D8 A0A069DZT4 A0A2A3EBU1 A0A2S2QJQ0 A0A154PCK7 A0A146M0N2 X1WW89 A0A125RM75 A0A1I8PGD2 A0A1I8M696 A0A1I8PGK1 A0A1I8M6A3 A0A1I8PGC3 A0A1I8PGJ1 A0A1I8M6A1 A0A1I8M699 A0A0Q9XGP2 A0A1I8M694 A0A1I8M691 A0A1I8M687 A0A1I8M6A2 A0A1I8PGK7 A0A1I8PGL6 T1PG89 A0A034V5U5 A0A1I8PGB6 T1PED2 A0A1I8PGJ8 A0A1I8PGC8 A0A1I8M698 A0A1I8PGB1 A0A1I8PGB4 A0A1I8PGJ2 A0A1I8M6A6 A0A034V2K4 A0A1I8M6B5 A0A1I8M692 A0A336KSQ1 A0A034V6X7 A0A0Q9X3T7 A0A0Q9XDD3 A0A0Q9XEN4 A0A0Q9WL68 A0A1B0F930 A0A1I8PGK2 A0A1I8PGB9 A0A0Q9X475 N6UIL0 A0A0Q9WLL0 E2B8D1 A0A0Q9X3Q7 A0A0Q9XGA4 A0A336MD69 A0A1I8PGL1 A0A1I8M6A8 A0A1I8M6A4 A0A1I8M6B3 A0A0Q9WXE7 A0A0Q9WYS7 A0A1I8M6B0 A0A0Q9WWH8 A0A0Q9WLR3 A0A1I8PGC2 A0A0Q9WMZ8 A0A336KWU5 A0A1I8PGK6 A0A1I8M6B1 A0A1I8PGJ6 A0A0Q9X4D4 A0A1I8PGK3 B4KIM7 A0A1I8M6A9 A0A1Q3FWN9 A0A336MEP5 A0A0R3P0Y9
Pubmed
EMBL
KY484778
ASS36020.1
KM453742
KP335687
AKC34057.1
AKC34899.1
+ More
KY796050 ARE31046.1 GU357490 ADB45217.1 KP335686 KM244042 AKC42335.1 RSAL01000139 RVE46159.1 KK107390 EZA51426.1 KQ971371 KYB25508.1 GEZM01012123 JAV93174.1 GEZM01012122 JAV93175.1 GFDF01003928 JAV10156.1 JXUM01023791 JXUM01023792 JXUM01023793 KQ560671 KXJ81341.1 GEDC01001842 GEDC01000137 JAS35456.1 JAS37161.1 GEDC01025119 GEDC01005564 GEDC01003693 JAS12179.1 JAS31734.1 JAS33605.1 GFDL01003050 JAV31995.1 KQ435944 KOX68195.1 CVRI01000054 CRL00440.1 KF828785 AIN44102.1 GBBI01002685 JAC16027.1 GBGD01000085 JAC88804.1 KZ288292 PBC29188.1 GGMS01008735 MBY77938.1 KQ434864 KZC08998.1 GDHC01006312 JAQ12317.1 ABLF02016184 ABLF02016186 KT428603 AMD40296.1 CH933807 KRG02659.1 KA647155 AFP61784.1 GAKP01021440 JAC37512.1 KA647049 AFP61678.1 GAKP01021441 JAC37511.1 UFQS01000890 UFQT01000890 SSX07519.1 SSX27859.1 GAKP01021442 JAC37510.1 KRG02667.1 KRG02658.1 KRG02657.1 CH940661 KRF85612.1 KRG02669.1 APGK01023890 KB740523 ENN80486.1 KRF85625.1 GL446310 EFN88051.1 KRG02662.1 KRG02664.1 SSX07520.1 SSX27860.1 KRF85613.1 KRF85627.1 KRF85631.1 KRF85621.1 KRF85615.1 KRF85619.1 KRF85622.1 UFQS01000910 UFQT01000910 SSX07613.1 SSX27951.1 KRG02661.1 EDW11370.2 GFDL01003048 JAV31997.1 SSX27861.1 CH379061 KRT04598.1
KY796050 ARE31046.1 GU357490 ADB45217.1 KP335686 KM244042 AKC42335.1 RSAL01000139 RVE46159.1 KK107390 EZA51426.1 KQ971371 KYB25508.1 GEZM01012123 JAV93174.1 GEZM01012122 JAV93175.1 GFDF01003928 JAV10156.1 JXUM01023791 JXUM01023792 JXUM01023793 KQ560671 KXJ81341.1 GEDC01001842 GEDC01000137 JAS35456.1 JAS37161.1 GEDC01025119 GEDC01005564 GEDC01003693 JAS12179.1 JAS31734.1 JAS33605.1 GFDL01003050 JAV31995.1 KQ435944 KOX68195.1 CVRI01000054 CRL00440.1 KF828785 AIN44102.1 GBBI01002685 JAC16027.1 GBGD01000085 JAC88804.1 KZ288292 PBC29188.1 GGMS01008735 MBY77938.1 KQ434864 KZC08998.1 GDHC01006312 JAQ12317.1 ABLF02016184 ABLF02016186 KT428603 AMD40296.1 CH933807 KRG02659.1 KA647155 AFP61784.1 GAKP01021440 JAC37512.1 KA647049 AFP61678.1 GAKP01021441 JAC37511.1 UFQS01000890 UFQT01000890 SSX07519.1 SSX27859.1 GAKP01021442 JAC37510.1 KRG02667.1 KRG02658.1 KRG02657.1 CH940661 KRF85612.1 KRG02669.1 APGK01023890 KB740523 ENN80486.1 KRF85625.1 GL446310 EFN88051.1 KRG02662.1 KRG02664.1 SSX07520.1 SSX27860.1 KRF85613.1 KRF85627.1 KRF85631.1 KRF85621.1 KRF85615.1 KRF85619.1 KRF85622.1 UFQS01000910 UFQT01000910 SSX07613.1 SSX27951.1 KRG02661.1 EDW11370.2 GFDL01003048 JAV31997.1 SSX27861.1 CH379061 KRT04598.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A2H4KAX1
A0A0F6PP97
A0A220A455
D2Y5V6
A0A0F6PQJ9
A0A3S2NW63
+ More
K7J1C7 A0A026W727 A0A139WC08 A0A1W4XSV3 A0A1Y1N5M9 A0A1Y1N973 A0A1W4XTX1 A0A1L8DUP5 A0A1W4XTS4 A0A182H2B8 A0A1W4XIR5 A0A1B6EGY9 A0A1B6E1F3 A0A1Q3FWR2 A0A1S4F8S7 A0A0M8ZP27 A0A1J1IJS3 A0A161ANL7 A0A023F4D8 A0A069DZT4 A0A2A3EBU1 A0A2S2QJQ0 A0A154PCK7 A0A146M0N2 X1WW89 A0A125RM75 A0A1I8PGD2 A0A1I8M696 A0A1I8PGK1 A0A1I8M6A3 A0A1I8PGC3 A0A1I8PGJ1 A0A1I8M6A1 A0A1I8M699 A0A0Q9XGP2 A0A1I8M694 A0A1I8M691 A0A1I8M687 A0A1I8M6A2 A0A1I8PGK7 A0A1I8PGL6 T1PG89 A0A034V5U5 A0A1I8PGB6 T1PED2 A0A1I8PGJ8 A0A1I8PGC8 A0A1I8M698 A0A1I8PGB1 A0A1I8PGB4 A0A1I8PGJ2 A0A1I8M6A6 A0A034V2K4 A0A1I8M6B5 A0A1I8M692 A0A336KSQ1 A0A034V6X7 A0A0Q9X3T7 A0A0Q9XDD3 A0A0Q9XEN4 A0A0Q9WL68 A0A1B0F930 A0A1I8PGK2 A0A1I8PGB9 A0A0Q9X475 N6UIL0 A0A0Q9WLL0 E2B8D1 A0A0Q9X3Q7 A0A0Q9XGA4 A0A336MD69 A0A1I8PGL1 A0A1I8M6A8 A0A1I8M6A4 A0A1I8M6B3 A0A0Q9WXE7 A0A0Q9WYS7 A0A1I8M6B0 A0A0Q9WWH8 A0A0Q9WLR3 A0A1I8PGC2 A0A0Q9WMZ8 A0A336KWU5 A0A1I8PGK6 A0A1I8M6B1 A0A1I8PGJ6 A0A0Q9X4D4 A0A1I8PGK3 B4KIM7 A0A1I8M6A9 A0A1Q3FWN9 A0A336MEP5 A0A0R3P0Y9
K7J1C7 A0A026W727 A0A139WC08 A0A1W4XSV3 A0A1Y1N5M9 A0A1Y1N973 A0A1W4XTX1 A0A1L8DUP5 A0A1W4XTS4 A0A182H2B8 A0A1W4XIR5 A0A1B6EGY9 A0A1B6E1F3 A0A1Q3FWR2 A0A1S4F8S7 A0A0M8ZP27 A0A1J1IJS3 A0A161ANL7 A0A023F4D8 A0A069DZT4 A0A2A3EBU1 A0A2S2QJQ0 A0A154PCK7 A0A146M0N2 X1WW89 A0A125RM75 A0A1I8PGD2 A0A1I8M696 A0A1I8PGK1 A0A1I8M6A3 A0A1I8PGC3 A0A1I8PGJ1 A0A1I8M6A1 A0A1I8M699 A0A0Q9XGP2 A0A1I8M694 A0A1I8M691 A0A1I8M687 A0A1I8M6A2 A0A1I8PGK7 A0A1I8PGL6 T1PG89 A0A034V5U5 A0A1I8PGB6 T1PED2 A0A1I8PGJ8 A0A1I8PGC8 A0A1I8M698 A0A1I8PGB1 A0A1I8PGB4 A0A1I8PGJ2 A0A1I8M6A6 A0A034V2K4 A0A1I8M6B5 A0A1I8M692 A0A336KSQ1 A0A034V6X7 A0A0Q9X3T7 A0A0Q9XDD3 A0A0Q9XEN4 A0A0Q9WL68 A0A1B0F930 A0A1I8PGK2 A0A1I8PGB9 A0A0Q9X475 N6UIL0 A0A0Q9WLL0 E2B8D1 A0A0Q9X3Q7 A0A0Q9XGA4 A0A336MD69 A0A1I8PGL1 A0A1I8M6A8 A0A1I8M6A4 A0A1I8M6B3 A0A0Q9WXE7 A0A0Q9WYS7 A0A1I8M6B0 A0A0Q9WWH8 A0A0Q9WLR3 A0A1I8PGC2 A0A0Q9WMZ8 A0A336KWU5 A0A1I8PGK6 A0A1I8M6B1 A0A1I8PGJ6 A0A0Q9X4D4 A0A1I8PGK3 B4KIM7 A0A1I8M6A9 A0A1Q3FWN9 A0A336MEP5 A0A0R3P0Y9
PDB
6BHU
E-value=0,
Score=3678
Ontologies
GO
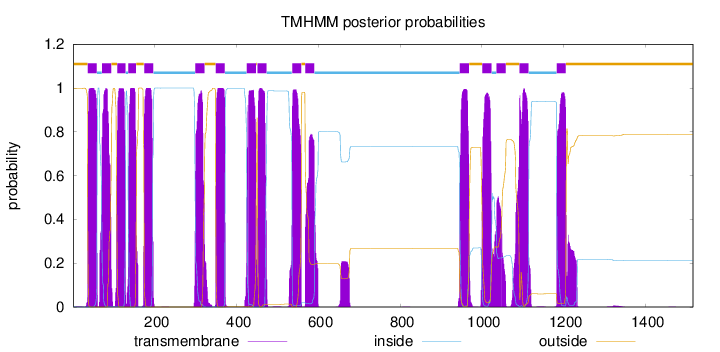
Topology
Length:
1516
Number of predicted TMHs:
16
Exp number of AAs in TMHs:
353.09155
Exp number, first 60 AAs:
22.73936
Total prob of N-in:
0.00168
POSSIBLE N-term signal
sequence
outside
1 - 35
TMhelix
36 - 58
inside
59 - 70
TMhelix
71 - 93
outside
94 - 107
TMhelix
108 - 128
inside
129 - 134
TMhelix
135 - 154
outside
155 - 173
TMhelix
174 - 196
inside
197 - 298
TMhelix
299 - 321
outside
322 - 348
TMhelix
349 - 371
inside
372 - 424
TMhelix
425 - 447
outside
448 - 450
TMhelix
451 - 473
inside
474 - 535
TMhelix
536 - 558
outside
559 - 567
TMhelix
568 - 590
inside
591 - 945
TMhelix
946 - 968
outside
969 - 1000
TMhelix
1001 - 1023
inside
1024 - 1035
TMhelix
1036 - 1058
outside
1059 - 1091
TMhelix
1092 - 1114
inside
1115 - 1182
TMhelix
1183 - 1205
outside
1206 - 1516
Population Genetic Test Statistics
Pi
17.608545
Theta
16.445644
Tajima's D
-1.675673
CLR
0.802215
CSRT
0.0378481075946203
Interpretation
Uncertain
