Gene
KWMTBOMO07382
Pre Gene Modal
BGIBMGA010332
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_multidrug_resistance-associated_protein_1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.735
Sequence
CDS
ATGACCATATACTTCCGGTTGCCGCAAACATTGGCTTTCCAACACATCATGAAAATATTGACGTTTACGTTGTCAGCAAGCTTAGTATGGCTTAACAAAAAACATGGAATGCGAGCTTCCGGCGTACTGTTTTTCTTCTGGTTACTGTTAGCTGTCATCGAAGTACCCAGGCTGTGGTGCAAAGCATTGGAATTGAAAATGAACTTCGTAAACGGAATACAATACGGATTTATTAGTCTTGCTATCCATTACGGACTGATAGTATTATTGTTAATATTGAATTGTTTCGCTGACGCAGCGCCACGTGATTCCGTGTACACGGATTCTCAGAACCAATGTCCCGAAAGATCTGCTGGTTTTCTAAGTCGTCTAACCTTCAGTTGGTTCGATTCTTTAGTACTAAAAGGATATCGTAAGGAATTTGAAGAAGGCGATGTGTGGGATTTGAATTATCAAGAATCTTCTTCAGAAGTCGTACCAAGATTTAAGAAATACTGGGAACAACCGGCAAATAGACGTAACTCATCAACAAAACACATAAAAGCAAAATTTCGAAGAAGATCAACTGATAGAACCATGAAAACTAATACTAAAAATCGAACTGTGTCAATATTACCAGCTCTTTGCTTTGCCTTCGGAGGACAGTTCATGTTTGGGACATTCCTGAAACTGCTGAGTGATATTTCCATTTTTTTATCACCACAACTGCTCAAGCTTCTTATAGGGTTTATAAGCAGCGATGAGCCAACATGGAGGGGCTACTTATATTCTTTCCTGTTCTTTGGTTGTGCCGCTCTTGAAACAACGCTGTTGAGTCATTACACTTTGAAAATGAACATTGTCGGAATGAGGATAAGAACAGCATTAACGAACGCTATTTACCAAATGGCTCTTCGCATGAACAATACGACGCGGAAGCAGTCGACAGTTGGTGAAATTATTAATCTGATGTCCGTGGATGCCCAAAGATTCGTCAGTTTGACAAGCACATTAAACATGATTTGGTCTGCGCCTTTTCAGATTGTGCTTGCTATTTATTTTTTGTGGGGCGTACTAGGTCCTTCTGTTCTCGCCGGATTGGCGGTTATGATTCTTTTAATTCCAATGAACGGTTTCATTGCTCGTATGGAAGAGAAACTGGAAATCAGATTAATGAAATTCAAAGATAACAGAATTAAAATCATGAATGAGGTGTTGAACGGAATTAAGGTCCTAAAAATGTATGCTTGGGAATTAAGTTTTCAAGGACAGATTGACCAAGTGAGAAATAAAGAACTTAATATACTGAAGAAAGTGATTTACTTAGATTCCCTGACGTCATTAACGTGGTCTGCGACACCATTTCTGGTTTCACTCACGTCTTTTGGGTGTTTTATTTTAGTAAATGATTCGGAGAGTTTGGATTCAAAGAAATTATTTGTGGCCTTATCTCTATTCAACATATTACGTTATCCATTAATGATGCTGCCAAACACAATATCAAATATCGTTCAAACTTCCGTTAGCATAAAACGTTTAAATAAATTTTTGAATTCAACGGTTTTAGATTTAGAATCAATCGAACATGACCCTCAATATTCCTATCCGATAACTGTGGAAAACGGACACTTCTCGTGGGAAGGTCAAGATTCTAGTACGGTGTTGAGAAATATAAATTTGCGTGTGCCCCGAGGTTCTTTGGTGGCAATAGTCGGTGCTGTGGGCGCCGGGAAGAGCTCGTTACTGTCCGCACTTTTGGGCGAGATGACTAAGGTGTCGGGACGTGTCAACGTTCACGGTACAATAGCATACGTGTCACAAGAGCCATGGATACAGAATGCCACATTGAAAGATAACATTCTCTTCGGGAAGCCGTTTAATAAATACAAATATAAGAATGTACTAAACGCCTGTACTCTTGTGCAAGACTTGGAAATATTTCCCGGAGGAGATCTAACTGAGATAGGTGAAAAAGGTATAAACTTATCGGGCGGTCAGAAGCAGCGCGTGTCGCTGGCGCGCGCCGTGTACCACGAGGCGGACAACTACCTGCTGGACGACCCGCTCAGCGCCGTGGACTCGCACGTCGGCAAGCACATCTTCGACAAGGTCATCGGGCCCGCCGGCCTGCTCAAGGACAGGACGCGCGTCTGGGTCACGCACAACGTGTCCTACCTCGCGCAGACCGACCTGGTCGTCGTGCTGCGCGACGGACAGGTCTCCGAGGCCGGCTCCTACCAGCACCTGCTCGAGAAGAAGGGCGCCTTTGCTGACTTCCTGCTTCATCATCTAAGCGATGCAGAACGGTCATCAGTCGAATCTGTCAGAAAACAAGACCACGAATACAGATGCTCAAGGACAGAATCCCACCAGAATTTACAAAGAATTCGTTTACATTCCGAGAGCAACTCAGAAGCAGAGGACCGATCAGGAAAACTAGCTATCAGCTTTAAAAGAAGTATCGAAGAAATTATTTTAAATAAATCCGCCGAGCACGTTAGTTATACTGAAAAAGAGAAATCTGAAAGGGGCAGTGTTTCGTGGATGGTTTACAAGCACTACTTGAAAAACATCGGCGTACTGGCCGCAGCAGCTACTATACTAATGAACGTGGTTCTACAGCTCTTTCATATCGGTTCGAGCTATTGGTTGGCAGAATGGTCCGACAACGAGAACGAAGTTGTTAACAGTACTATGAATTTGAACCAGCGGAACAAGTACCTCGCAGTATACGCTGGCCTCGGTATGGGACAAGCGATATCGTCATTTCTAGCCGACTTGATACCATACTTGGCGTGCTGGCGAGCTGCGAAGGTCATCCACGAACTGTTACTGGACAATGTGTTAAAGACGCCGTTGCATTTTTTCGAAGTCACGCCTATTGGCAGAATACTGTCAAGATTCTCTAAAGATATTGAAATTATTGACTCTATGCTACCTTCTCAATTAATCAATGGGGTGTATTGCGTCTTCGAACTCATTGGCATTTTATTCGTGATAAGCTTGTCGACGCCATCATTCTTAGCAATAGTTGTACCAGTAGGTCTAGTGTACTATGTAATACAACGGTTCTACGTGGCTACGTCTCGTCAGCTCAAGCGGCTAGAGTCCATAGCTCGCTCGCCGATATTCTCACACTTCGGCGAAACGATCAGTGGAGCAGCCACCATTCGCGCTTTCAACGTAACCGAGAGGTTTATTGAAGAATCAAATGAAAAGTTAGATCGCAACCTCGCTTATTGCTATGCGATTGTAGTAGGAAAGCGATGGCTTGGTTTCCGATTAGAATTCATTGGAAACTTCATATTATTTTTCGCTGCAATGTTTGCTGTTCTCAGCAGAGATACTATAAGCTCGGGTCTCGTGGGACTTTCCTTAACCTACGCTATGCAGGTAACTGCATCGTTGAATTGGCTTGTTCGGCTAGCTTCTGAAATTGAAAGTAATATTGTAGCTGTAGAACGGATAAAAGAATACTCTGAGAAAAATAAGGAAGCTGCGTGGAACTTAGGGAAAGGGCCCGGCGTGACGTGGCCCGAGACGGGAGCGCTACAGCTGGAGCAGCTGACGCTGGAGTACCGCCCCGGCGAGCCGGCGCTGCGTGACGTCACGTGCACGGTCGCGCCGCGCGACAAGCTCGGCATCGTGGGACGCACCGGCGCCGGCAAGTCTACTCTCACGCTCGGGCTCTTCAGGATCGTGGAGGCAGCCGCCGGACGCATCCTGATCGACGGCGTCGACATCGCGACCCTCGGGCTGCACCAGCTGCGGTCGCGCATCACCATCATCCCGCAGGAGCCGATCCTGTTCTCGGGCACGCTGCGCACCAACCTGGACCCGTTCGAGGCGTATAGCGACGAGGAGATCTGGCGCGCGCTGGAACACGCCCACCTCCGTGCCTTCGTGCAGGGGCTGCCGGCCGGGCTGCGGCACGAGGTGGCGGAGGGCGGCGAGAACCTGTCGGTGGGACAGCGCCAGCTCGTGTGCCTGGCGCGCGCGCTGCTACGCAAGACGCCGCTGCTGGTGCTGGACGAGGCCACCGCGGCCGTCGACCTCGAGACTGACGACCTCATCCAGAAGACGATCCGCTCGGAGTTCGCGTCGTGCACGGTGCTCACTATCGCGCACCGCCTCAACACCATCATGGACTCCACCAAAGTGATGGTGCTCGACCGCGGGCAGCTCGTAGAATATGCGGCCCCTCAACAACTACTTAACGATAAAAACTCCATTTTCTACTCCTTAGCAAAAGATGCTGGAATTGTCAGTTGA
Protein
MTIYFRLPQTLAFQHIMKILTFTLSASLVWLNKKHGMRASGVLFFFWLLLAVIEVPRLWCKALELKMNFVNGIQYGFISLAIHYGLIVLLLILNCFADAAPRDSVYTDSQNQCPERSAGFLSRLTFSWFDSLVLKGYRKEFEEGDVWDLNYQESSSEVVPRFKKYWEQPANRRNSSTKHIKAKFRRRSTDRTMKTNTKNRTVSILPALCFAFGGQFMFGTFLKLLSDISIFLSPQLLKLLIGFISSDEPTWRGYLYSFLFFGCAALETTLLSHYTLKMNIVGMRIRTALTNAIYQMALRMNNTTRKQSTVGEIINLMSVDAQRFVSLTSTLNMIWSAPFQIVLAIYFLWGVLGPSVLAGLAVMILLIPMNGFIARMEEKLEIRLMKFKDNRIKIMNEVLNGIKVLKMYAWELSFQGQIDQVRNKELNILKKVIYLDSLTSLTWSATPFLVSLTSFGCFILVNDSESLDSKKLFVALSLFNILRYPLMMLPNTISNIVQTSVSIKRLNKFLNSTVLDLESIEHDPQYSYPITVENGHFSWEGQDSSTVLRNINLRVPRGSLVAIVGAVGAGKSSLLSALLGEMTKVSGRVNVHGTIAYVSQEPWIQNATLKDNILFGKPFNKYKYKNVLNACTLVQDLEIFPGGDLTEIGEKGINLSGGQKQRVSLARAVYHEADNYLLDDPLSAVDSHVGKHIFDKVIGPAGLLKDRTRVWVTHNVSYLAQTDLVVVLRDGQVSEAGSYQHLLEKKGAFADFLLHHLSDAERSSVESVRKQDHEYRCSRTESHQNLQRIRLHSESNSEAEDRSGKLAISFKRSIEEIILNKSAEHVSYTEKEKSERGSVSWMVYKHYLKNIGVLAAAATILMNVVLQLFHIGSSYWLAEWSDNENEVVNSTMNLNQRNKYLAVYAGLGMGQAISSFLADLIPYLACWRAAKVIHELLLDNVLKTPLHFFEVTPIGRILSRFSKDIEIIDSMLPSQLINGVYCVFELIGILFVISLSTPSFLAIVVPVGLVYYVIQRFYVATSRQLKRLESIARSPIFSHFGETISGAATIRAFNVTERFIEESNEKLDRNLAYCYAIVVGKRWLGFRLEFIGNFILFFAAMFAVLSRDTISSGLVGLSLTYAMQVTASLNWLVRLASEIESNIVAVERIKEYSEKNKEAAWNLGKGPGVTWPETGALQLEQLTLEYRPGEPALRDVTCTVAPRDKLGIVGRTGAGKSTLTLGLFRIVEAAAGRILIDGVDIATLGLHQLRSRITIIPQEPILFSGTLRTNLDPFEAYSDEEIWRALEHAHLRAFVQGLPAGLRHEVAEGGENLSVGQRQLVCLARALLRKTPLLVLDEATAAVDLETDDLIQKTIRSEFASCTVLTIAHRLNTIMDSTKVMVLDRGQLVEYAAPQQLLNDKNSIFYSLAKDAGIVS
Summary
Uniprot
A0A220A455
A0A2H4KAX1
A0A0F6PP97
D2Y5V6
A0A0F6PQJ9
A0A1W4XTX1
+ More
A0A158NRB1 A0A1B6E1F3 A0A1B6EGY9 A0A139WC08 A0A1Y1N5M9 A0A1W4XSV3 A0A067RTQ5 A0A1W4XIR5 A0A182H2B8 A0A1Y1N973 E2B8D1 A0A0Q9X475 A0A1J1IJS3 A0A1L8DUP5 K7J1C7 A0A0M8ZP27 A0A026W727 A0A0Q9WLS5 A0A1I8PGD2 A0A0Q9WLR3 A0A1W4XTS4 A0A2A3EBU1 A0A0Q9WVA8 A0A069DZT4 A0A023F4D8 A0A1I8PGJ8 A0A1I8PGB1 A0A1I8M6A2 A0A0Q9XGP2 A0A0Q9XB26 A0A224X9P4 A0A0Q9WYS7 A0A0R3NUI1 A0A0Q5WA21 A0A0Q9XDD3 A0A1I8M6A1 A0A1I8M699 A0A1Q3FWR2 A0A1I8M694 A0A0Q9WLL0 A0A0Q9WL68 A0A0P9BPP2 A0A0Q9WXE7 A0A034V6X7 X1WW89 A0A1I8M6A6 A0A1I8M6B5 A0A0Q9X3T7 A0A0Q9WX91 A0A0Q9XEN4 A0A0Q9WMZ8 Q7KTB7 A0A125RM75 A0A0J9R1I1 A0A336KSQ1 A0A154PCK7 A0A0Q9WWH8 A0A1I8PGJ1 A0A1I8PGK1 A0A1I8PGC3 A0A1I8PGB4 A0A1I8PGB6 A0A1W4V5U0 A0A1I8PGL6 A0A3R5WTB4 A0A146M0N2 A0A1I8PGJ2 A0A1I8PGC8 A0A1I8PGK7 A0A336MD69 Q29L70 A0A0R1DK17 A0A1Q3FWN9 A0A0Q9XGA4 A0A1I8M6B3 A0A0Q9X3Q7 A0A1S4F8S7 A0A161ANL7 A0A1I8M698 V5I844 A0A1I8M691 A0A1I8M692 A0A1I8M687 A0A0R3P0Y9 A0A1L8DUK0 A0A2S2QJQ0 A0A336KWU5 B4KIM7 A0A0Q9WL82 A0A0P8XGE2 A0A0Q9WLJ8 T1PED2 A0A0Q9WXA1 T1PG89
A0A158NRB1 A0A1B6E1F3 A0A1B6EGY9 A0A139WC08 A0A1Y1N5M9 A0A1W4XSV3 A0A067RTQ5 A0A1W4XIR5 A0A182H2B8 A0A1Y1N973 E2B8D1 A0A0Q9X475 A0A1J1IJS3 A0A1L8DUP5 K7J1C7 A0A0M8ZP27 A0A026W727 A0A0Q9WLS5 A0A1I8PGD2 A0A0Q9WLR3 A0A1W4XTS4 A0A2A3EBU1 A0A0Q9WVA8 A0A069DZT4 A0A023F4D8 A0A1I8PGJ8 A0A1I8PGB1 A0A1I8M6A2 A0A0Q9XGP2 A0A0Q9XB26 A0A224X9P4 A0A0Q9WYS7 A0A0R3NUI1 A0A0Q5WA21 A0A0Q9XDD3 A0A1I8M6A1 A0A1I8M699 A0A1Q3FWR2 A0A1I8M694 A0A0Q9WLL0 A0A0Q9WL68 A0A0P9BPP2 A0A0Q9WXE7 A0A034V6X7 X1WW89 A0A1I8M6A6 A0A1I8M6B5 A0A0Q9X3T7 A0A0Q9WX91 A0A0Q9XEN4 A0A0Q9WMZ8 Q7KTB7 A0A125RM75 A0A0J9R1I1 A0A336KSQ1 A0A154PCK7 A0A0Q9WWH8 A0A1I8PGJ1 A0A1I8PGK1 A0A1I8PGC3 A0A1I8PGB4 A0A1I8PGB6 A0A1W4V5U0 A0A1I8PGL6 A0A3R5WTB4 A0A146M0N2 A0A1I8PGJ2 A0A1I8PGC8 A0A1I8PGK7 A0A336MD69 Q29L70 A0A0R1DK17 A0A1Q3FWN9 A0A0Q9XGA4 A0A1I8M6B3 A0A0Q9X3Q7 A0A1S4F8S7 A0A161ANL7 A0A1I8M698 V5I844 A0A1I8M691 A0A1I8M692 A0A1I8M687 A0A0R3P0Y9 A0A1L8DUK0 A0A2S2QJQ0 A0A336KWU5 B4KIM7 A0A0Q9WL82 A0A0P8XGE2 A0A0Q9WLJ8 T1PED2 A0A0Q9WXA1 T1PG89
Pubmed
EMBL
KY796050
ARE31046.1
KY484778
ASS36020.1
KM453742
KP335687
+ More
AKC34057.1 AKC34899.1 GU357490 ADB45217.1 KP335686 KM244042 AKC42335.1 ADTU01023843 ADTU01023844 GEDC01025119 GEDC01005564 GEDC01003693 JAS12179.1 JAS31734.1 JAS33605.1 GEDC01001842 GEDC01000137 JAS35456.1 JAS37161.1 KQ971371 KYB25508.1 GEZM01012123 JAV93174.1 KK852424 KDR24195.1 JXUM01023791 JXUM01023792 JXUM01023793 KQ560671 KXJ81341.1 GEZM01012122 JAV93175.1 GL446310 EFN88051.1 CH933807 KRG02669.1 CVRI01000054 CRL00440.1 GFDF01003928 JAV10156.1 KQ435944 KOX68195.1 KK107390 EZA51426.1 CH940661 KRF85626.1 KRF85619.1 KZ288292 PBC29188.1 KRF85617.1 GBGD01000085 JAC88804.1 GBBI01002685 JAC16027.1 KRG02659.1 KRG02666.1 GFTR01008802 JAW07624.1 KRF85621.1 CH379061 KRT04588.1 CH954177 KQS70503.1 KRG02658.1 GFDL01003050 JAV31995.1 KRF85625.1 KRF85612.1 CH902620 KPU73781.1 KRF85613.1 KRF85627.1 KRF85631.1 GAKP01021442 JAC37510.1 ABLF02016184 ABLF02016186 KRG02667.1 KRF85614.1 KRG02657.1 KRF85622.1 AE014134 AAS64699.1 KT428603 AMD40296.1 CM002910 KMY89971.1 UFQS01000890 UFQT01000890 SSX07519.1 SSX27859.1 KQ434864 KZC08998.1 KRF85615.1 MH172499 QAA95906.1 GDHC01006312 JAQ12317.1 SSX07520.1 SSX27860.1 EAL32954.3 CM000157 KRJ97652.1 KRJ97659.1 GFDL01003048 JAV31997.1 KRG02664.1 KRG02662.1 KF828785 AIN44102.1 GALX01005330 JAB63136.1 KRT04598.1 GFDF01003977 JAV10107.1 GGMS01008735 MBY77938.1 UFQS01000910 UFQT01000910 SSX07613.1 SSX27951.1 EDW11370.2 KRF85628.1 KPU73769.1 KRF85624.1 KA647049 AFP61678.1 KRF85630.1 KA647155 AFP61784.1
AKC34057.1 AKC34899.1 GU357490 ADB45217.1 KP335686 KM244042 AKC42335.1 ADTU01023843 ADTU01023844 GEDC01025119 GEDC01005564 GEDC01003693 JAS12179.1 JAS31734.1 JAS33605.1 GEDC01001842 GEDC01000137 JAS35456.1 JAS37161.1 KQ971371 KYB25508.1 GEZM01012123 JAV93174.1 KK852424 KDR24195.1 JXUM01023791 JXUM01023792 JXUM01023793 KQ560671 KXJ81341.1 GEZM01012122 JAV93175.1 GL446310 EFN88051.1 CH933807 KRG02669.1 CVRI01000054 CRL00440.1 GFDF01003928 JAV10156.1 KQ435944 KOX68195.1 KK107390 EZA51426.1 CH940661 KRF85626.1 KRF85619.1 KZ288292 PBC29188.1 KRF85617.1 GBGD01000085 JAC88804.1 GBBI01002685 JAC16027.1 KRG02659.1 KRG02666.1 GFTR01008802 JAW07624.1 KRF85621.1 CH379061 KRT04588.1 CH954177 KQS70503.1 KRG02658.1 GFDL01003050 JAV31995.1 KRF85625.1 KRF85612.1 CH902620 KPU73781.1 KRF85613.1 KRF85627.1 KRF85631.1 GAKP01021442 JAC37510.1 ABLF02016184 ABLF02016186 KRG02667.1 KRF85614.1 KRG02657.1 KRF85622.1 AE014134 AAS64699.1 KT428603 AMD40296.1 CM002910 KMY89971.1 UFQS01000890 UFQT01000890 SSX07519.1 SSX27859.1 KQ434864 KZC08998.1 KRF85615.1 MH172499 QAA95906.1 GDHC01006312 JAQ12317.1 SSX07520.1 SSX27860.1 EAL32954.3 CM000157 KRJ97652.1 KRJ97659.1 GFDL01003048 JAV31997.1 KRG02664.1 KRG02662.1 KF828785 AIN44102.1 GALX01005330 JAB63136.1 KRT04598.1 GFDF01003977 JAV10107.1 GGMS01008735 MBY77938.1 UFQS01000910 UFQT01000910 SSX07613.1 SSX27951.1 EDW11370.2 KRF85628.1 KPU73769.1 KRF85624.1 KA647049 AFP61678.1 KRF85630.1 KA647155 AFP61784.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A220A455
A0A2H4KAX1
A0A0F6PP97
D2Y5V6
A0A0F6PQJ9
A0A1W4XTX1
+ More
A0A158NRB1 A0A1B6E1F3 A0A1B6EGY9 A0A139WC08 A0A1Y1N5M9 A0A1W4XSV3 A0A067RTQ5 A0A1W4XIR5 A0A182H2B8 A0A1Y1N973 E2B8D1 A0A0Q9X475 A0A1J1IJS3 A0A1L8DUP5 K7J1C7 A0A0M8ZP27 A0A026W727 A0A0Q9WLS5 A0A1I8PGD2 A0A0Q9WLR3 A0A1W4XTS4 A0A2A3EBU1 A0A0Q9WVA8 A0A069DZT4 A0A023F4D8 A0A1I8PGJ8 A0A1I8PGB1 A0A1I8M6A2 A0A0Q9XGP2 A0A0Q9XB26 A0A224X9P4 A0A0Q9WYS7 A0A0R3NUI1 A0A0Q5WA21 A0A0Q9XDD3 A0A1I8M6A1 A0A1I8M699 A0A1Q3FWR2 A0A1I8M694 A0A0Q9WLL0 A0A0Q9WL68 A0A0P9BPP2 A0A0Q9WXE7 A0A034V6X7 X1WW89 A0A1I8M6A6 A0A1I8M6B5 A0A0Q9X3T7 A0A0Q9WX91 A0A0Q9XEN4 A0A0Q9WMZ8 Q7KTB7 A0A125RM75 A0A0J9R1I1 A0A336KSQ1 A0A154PCK7 A0A0Q9WWH8 A0A1I8PGJ1 A0A1I8PGK1 A0A1I8PGC3 A0A1I8PGB4 A0A1I8PGB6 A0A1W4V5U0 A0A1I8PGL6 A0A3R5WTB4 A0A146M0N2 A0A1I8PGJ2 A0A1I8PGC8 A0A1I8PGK7 A0A336MD69 Q29L70 A0A0R1DK17 A0A1Q3FWN9 A0A0Q9XGA4 A0A1I8M6B3 A0A0Q9X3Q7 A0A1S4F8S7 A0A161ANL7 A0A1I8M698 V5I844 A0A1I8M691 A0A1I8M692 A0A1I8M687 A0A0R3P0Y9 A0A1L8DUK0 A0A2S2QJQ0 A0A336KWU5 B4KIM7 A0A0Q9WL82 A0A0P8XGE2 A0A0Q9WLJ8 T1PED2 A0A0Q9WXA1 T1PG89
A0A158NRB1 A0A1B6E1F3 A0A1B6EGY9 A0A139WC08 A0A1Y1N5M9 A0A1W4XSV3 A0A067RTQ5 A0A1W4XIR5 A0A182H2B8 A0A1Y1N973 E2B8D1 A0A0Q9X475 A0A1J1IJS3 A0A1L8DUP5 K7J1C7 A0A0M8ZP27 A0A026W727 A0A0Q9WLS5 A0A1I8PGD2 A0A0Q9WLR3 A0A1W4XTS4 A0A2A3EBU1 A0A0Q9WVA8 A0A069DZT4 A0A023F4D8 A0A1I8PGJ8 A0A1I8PGB1 A0A1I8M6A2 A0A0Q9XGP2 A0A0Q9XB26 A0A224X9P4 A0A0Q9WYS7 A0A0R3NUI1 A0A0Q5WA21 A0A0Q9XDD3 A0A1I8M6A1 A0A1I8M699 A0A1Q3FWR2 A0A1I8M694 A0A0Q9WLL0 A0A0Q9WL68 A0A0P9BPP2 A0A0Q9WXE7 A0A034V6X7 X1WW89 A0A1I8M6A6 A0A1I8M6B5 A0A0Q9X3T7 A0A0Q9WX91 A0A0Q9XEN4 A0A0Q9WMZ8 Q7KTB7 A0A125RM75 A0A0J9R1I1 A0A336KSQ1 A0A154PCK7 A0A0Q9WWH8 A0A1I8PGJ1 A0A1I8PGK1 A0A1I8PGC3 A0A1I8PGB4 A0A1I8PGB6 A0A1W4V5U0 A0A1I8PGL6 A0A3R5WTB4 A0A146M0N2 A0A1I8PGJ2 A0A1I8PGC8 A0A1I8PGK7 A0A336MD69 Q29L70 A0A0R1DK17 A0A1Q3FWN9 A0A0Q9XGA4 A0A1I8M6B3 A0A0Q9X3Q7 A0A1S4F8S7 A0A161ANL7 A0A1I8M698 V5I844 A0A1I8M691 A0A1I8M692 A0A1I8M687 A0A0R3P0Y9 A0A1L8DUK0 A0A2S2QJQ0 A0A336KWU5 B4KIM7 A0A0Q9WL82 A0A0P8XGE2 A0A0Q9WLJ8 T1PED2 A0A0Q9WXA1 T1PG89
PDB
6BHU
E-value=0,
Score=3277
Ontologies
GO
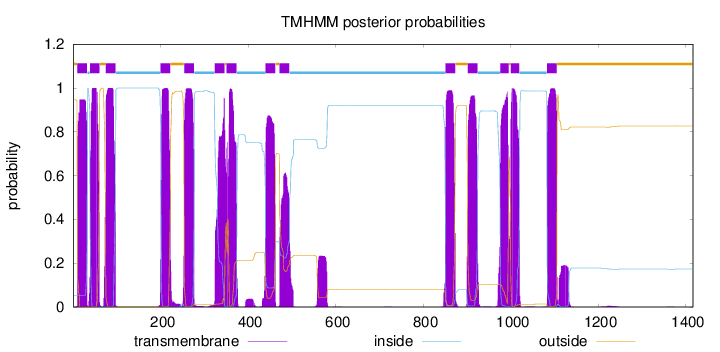
Topology
Length:
1416
Number of predicted TMHs:
14
Exp number of AAs in TMHs:
295.0844
Exp number, first 60 AAs:
40.8949
Total prob of N-in:
0.05342
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 32
inside
33 - 38
TMhelix
39 - 60
outside
61 - 74
TMhelix
75 - 97
inside
98 - 199
TMhelix
200 - 222
outside
223 - 253
TMhelix
254 - 276
inside
277 - 323
TMhelix
324 - 346
outside
347 - 350
TMhelix
351 - 373
inside
374 - 439
TMhelix
440 - 462
outside
463 - 471
TMhelix
472 - 494
inside
495 - 850
TMhelix
851 - 873
outside
874 - 901
TMhelix
902 - 924
inside
925 - 975
TMhelix
976 - 995
outside
996 - 999
TMhelix
1000 - 1019
inside
1020 - 1082
TMhelix
1083 - 1105
outside
1106 - 1416
Population Genetic Test Statistics
Pi
19.311822
Theta
17.929452
Tajima's D
-1.004854
CLR
0.637045
CSRT
0.131893405329734
Interpretation
Uncertain
