Gene
KWMTBOMO07378 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004955
Annotation
serine_protease_inhibitor_13_precursor_[Bombyx_mori]
Full name
Plasminogen activator inhibitor 1
Alternative Name
Endothelial plasminogen activator inhibitor
Serpin E1
Serpin E1
Location in the cell
Extracellular Reliability : 1.772
Sequence
CDS
ATGCGACGAAAAATCCAATTCTTTCTGTGCTGTAAACTTATCATCGGAACATGGTGTAACACTATGGACGGAGTAACAACAAAAGCAAATTCCTCAGAGGCTCAATTTCAACCAAATTATTTGGCAGACTCAGTCAACAATTTTGGGTACAAATTACTTTTAAAAATGATGGAGCAAAACAAGAATGAAAACATCGCATTGTCTCCGACCGGTATTGCTGGATTACTGGCAATGACACTTCTCGGTAGTGTGGGAAGTACATACGACGAGCTCGCCACAAGCCTCGGTTTTTCTCAAGACATATTGGCCAATCGAAACCACCACGAGCAGTTCGGTGAGCTGCTTCAACAATTGAACGATAATGAAACGAACTCCAAAACATTGTATGCAGATGCCATGTTTGTTGATTCACAAACCCGGATCCGGTCCGTTTTTAAAGATTATTTGACGACCGTGTACCACGGGGAAGCCCGCGGTGTCAACTTCACACAAAAGAATGAAGTCAAACAAATGATAAATGAGTGGGTCAAAGCGCAAACGCAGGCAAAGATTGAAGATTTTTTAAAACAATCACTACCAGAGGCAACAAAGGTGGTATTGCTAAGCGCTCTCTACTTCAGCGGACAATGGGCATCACCCTTTATTCCGGAATACACATTTAAGATGCCGTTTAAGAAACCTGAGGGCGAAGTAATGGCCGACCTTATGCTTAATTTAGGAGACTTCGATTACATTTTCTCTGAAAAAGACGGTTTGCATATGATCGCACTACCGTACAACGACTCTACAACCGTTATGTACGCTCTCAAACCTAGATTACCAATGAAACTAAGCCTTTTTGATCTGATGAAAAAACTAGACTACAATAAGATTGACGAACTAATTAATAAAATGAACGAAAGAAAGGCTGTTGTGCGATTCCCTAAAATGGACTTGGAGAGCAACGTTAATCTAGACAAAGCACTGAAAGCTGTCGGAATCAAGTCTATGTTCAGTCCCGACGAAGCTAATTTTGCTTTGATGATAGATTCGTCTGATGATAAAAAAGAAAACGAGGTAGTTATTAGATCGAGATTCGGTCACGAGCAGTCAAGAATGTTAAAAGACGCATTGAACAGTTTACCCAATCCGGGAGTGTACGTCGACTCCGTTTTACATAACGTCAAGATTACTGTTAACGAATTCGGAACGGAAGCTGTGGCGGCGACGGCCGGGATATTGGCGCGATCAGCTGAACAGTTCTACGCGGACTCCCCGTTCTACATATTCATTCGGAACGAGAAGACCAAACTAGTGACCTTTAGTGCCGTCGTATACGATCCTACGATATAG
Protein
MRRKIQFFLCCKLIIGTWCNTMDGVTTKANSSEAQFQPNYLADSVNNFGYKLLLKMMEQNKNENIALSPTGIAGLLAMTLLGSVGSTYDELATSLGFSQDILANRNHHEQFGELLQQLNDNETNSKTLYADAMFVDSQTRIRSVFKDYLTTVYHGEARGVNFTQKNEVKQMINEWVKAQTQAKIEDFLKQSLPEATKVVLLSALYFSGQWASPFIPEYTFKMPFKKPEGEVMADLMLNLGDFDYIFSEKDGLHMIALPYNDSTTVMYALKPRLPMKLSLFDLMKKLDYNKIDELINKMNERKAVVRFPKMDLESNVNLDKALKAVGIKSMFSPDEANFALMIDSSDDKKENEVVIRSRFGHEQSRMLKDALNSLPNPGVYVDSVLHNVKITVNEFGTEAVAATAGILARSAEQFYADSPFYIFIRNEKTKLVTFSAVVYDPTI
Summary
Description
Serine protease inhibitor. Inhibits TMPRSS7. Is a primary inhibitor of tissue-type plasminogen activator (PLAT) and urokinase-type plasminogen activator (PLAU). As PLAT inhibitor, it is required for fibrinolysis down-regulation and is responsible for the controlled degradation of blood clots. As PLAU inhibitor, it is involved in the regulation of cell adhesion and spreading. Acts as a regulator of cell migration, independently of its role as protease inhibitor. It is required for stimulation of keratinocyte migration during cutaneous injury repair. It is involved in cellular and replicative senescence (By similarity). Plays a role in alveolar type 2 cells senescence in the lung (By similarity). Is involved in the regulation of cementogenic differentiation of periodontal ligament stem cells, and regulates odontoblast differentiation and dentin formation during odontogenesis (By similarity).
Subunit
Forms a heterodimer with TMPRSS7. Interacts with VTN. Binds LRP1B; binding is followed by internalization and degradation. Interacts with PPP1CB.
Similarity
Belongs to the serpin family.
Keywords
Complete proteome
Glycoprotein
Protease inhibitor
Reference proteome
Secreted
Serine protease inhibitor
Signal
Feature
chain Plasminogen activator inhibitor 1
Uniprot
C0J8G2
A0A3G1VCH2
A0A2A4J5M2
A0A212F3H3
A0A194PVI0
A0A194R3B9
+ More
J7HLS7 S4NYK9 A0A2H1VQ07 X2J109 A0A1W4X1P5 A0A195F1F1 A0A2J7RSS5 A0A067R8V4 D2A2J9 A0A151XC37 J3JUV5 N6UPE2 U4UQY2 A0A1B6GCU3 A0A195CH67 A0A1B6JCM9 A0A026WZD2 A0A1V1FIT8 T1DGH8 T1DGG7 A0A084WQL3 Q16S06 P79335 B4MVS2 A0A1Q3FIJ5 U6CX60 G9KN61 A0A1S4JWS5 B0WX63 A0A023ESX4 A0A182J1Y2 P50449 A0A023ESG3 A0A2Y9E5Y9 A0A182H582 B3MJQ6 G3T7G7 B4KGA6 A0A023EV90 A0A182GK96 A0A2K6FA88 B4QBV3 I3LQQ6 A0A2Y9PNI9 A0A1W4V0X4 B4HNC1 H0UTC2 A0A3B0JGQ0 A0A286XDM0 C1JFL8 A0A0M3QT24 A0A1S2ZE36 A0A1B6DSP6 A0A0D9S037 S7Q4B4 A0A2M3ZZQ8 A0A2D0PLI2 A0A1Z4NK21 A0A252DWU7 A0A252DGG5 A0A1L8H381 A0A2M3ZZT6 A0A3L7HLU5 A0A1U7R3V0 K7FMQ9 A0A2K6TSH2 Q0IHE4 B4NWB7 G5E899 G5B5Z9 A0A2M4CUF6 B4JAP4 A0A2M3ZE19 A0A2M4BUU6 A0A1W4VWE9 G1NYA4 A0A182F4A5 A0A2M4BM55 A5D8P7 B4G7D2 A0A2M4AK75 A0A2M3ZFW7 Q29LV3
J7HLS7 S4NYK9 A0A2H1VQ07 X2J109 A0A1W4X1P5 A0A195F1F1 A0A2J7RSS5 A0A067R8V4 D2A2J9 A0A151XC37 J3JUV5 N6UPE2 U4UQY2 A0A1B6GCU3 A0A195CH67 A0A1B6JCM9 A0A026WZD2 A0A1V1FIT8 T1DGH8 T1DGG7 A0A084WQL3 Q16S06 P79335 B4MVS2 A0A1Q3FIJ5 U6CX60 G9KN61 A0A1S4JWS5 B0WX63 A0A023ESX4 A0A182J1Y2 P50449 A0A023ESG3 A0A2Y9E5Y9 A0A182H582 B3MJQ6 G3T7G7 B4KGA6 A0A023EV90 A0A182GK96 A0A2K6FA88 B4QBV3 I3LQQ6 A0A2Y9PNI9 A0A1W4V0X4 B4HNC1 H0UTC2 A0A3B0JGQ0 A0A286XDM0 C1JFL8 A0A0M3QT24 A0A1S2ZE36 A0A1B6DSP6 A0A0D9S037 S7Q4B4 A0A2M3ZZQ8 A0A2D0PLI2 A0A1Z4NK21 A0A252DWU7 A0A252DGG5 A0A1L8H381 A0A2M3ZZT6 A0A3L7HLU5 A0A1U7R3V0 K7FMQ9 A0A2K6TSH2 Q0IHE4 B4NWB7 G5E899 G5B5Z9 A0A2M4CUF6 B4JAP4 A0A2M3ZE19 A0A2M4BUU6 A0A1W4VWE9 G1NYA4 A0A182F4A5 A0A2M4BM55 A5D8P7 B4G7D2 A0A2M4AK75 A0A2M3ZFW7 Q29LV3
Pubmed
EMBL
EU935614
ACG61176.1
MG732911
AYK02793.1
NWSH01003223
PCG66730.1
+ More
AGBW02010564 OWR48286.1 KQ459592 KPI96769.1 KQ460845 KPJ12024.1 JN400446 AFQ01145.1 GAIX01008519 JAA84041.1 ODYU01003476 SOQ42314.1 KJ127528 AHN49718.1 KQ981864 KYN34293.1 NEVH01000250 PNF43885.1 KK852808 KDR16030.1 KQ971338 EFA02869.2 KQ982314 KYQ57956.1 BT127020 AEE61982.1 APGK01023288 APGK01023289 APGK01023290 KB740481 ENN80572.1 KB632431 ERL95517.1 GECZ01009507 JAS60262.1 KQ977754 KYN00081.1 GECU01027274 GECU01010869 JAS80432.1 JAS96837.1 KK107063 EZA61168.1 FX985318 BAX07331.1 GALA01001651 JAA93201.1 GALA01001666 JAA93186.1 ATLV01025637 KE525396 KFB52507.1 CH477692 EAT37202.1 Y11347 CH963857 EDW75792.2 GFDL01007624 JAV27421.1 HAAF01002611 CCP74437.1 AEYP01053153 JP017742 AES06340.1 DS232158 EDS36324.1 GAPW01001200 JAC12398.1 X58541 GAPW01001393 JAC12205.1 JXUM01026061 KQ560742 KXJ81047.1 CH902620 EDV31395.2 CH933807 EDW11093.1 GAPW01001199 JAC12399.1 JXUM01069686 JXUM01069687 JXUM01069688 JXUM01069689 KQ562564 KXJ75606.1 CM000362 CM002911 EDX07613.1 KMY94760.1 AEMK02000015 CH480816 EDW48403.1 AAKN02032075 OUUW01000004 SPP79442.1 FJ608558 ACO24971.1 CP012523 ALC38139.1 GEDC01008592 JAS28706.1 AQIB01127710 KE164168 EPQ15712.1 GGFK01000735 MBW34056.1 AP018254 BAZ06050.1 MTAW01000089 OUL32095.1 MTAX01000054 OUL26509.1 CM004470 OCT90552.1 GGFK01000742 MBW34063.1 RAZU01000172 RLQ66934.1 AGCU01051555 BC123188 AAI23189.1 CM000157 EDW88434.1 AC147986 CH466529 EDL19287.1 JH168609 EHB04710.1 GGFL01004633 MBW68811.1 CH916368 EDW03852.1 GGFM01006066 MBW26817.1 GGFJ01007696 MBW56837.1 AAPE02003478 GGFJ01005029 MBW54170.1 BC141762 AAI41763.1 CH479180 EDW29265.1 GGFK01007811 MBW41132.1 GGFM01006579 MBW27330.1 CH379060 EAL33942.2
AGBW02010564 OWR48286.1 KQ459592 KPI96769.1 KQ460845 KPJ12024.1 JN400446 AFQ01145.1 GAIX01008519 JAA84041.1 ODYU01003476 SOQ42314.1 KJ127528 AHN49718.1 KQ981864 KYN34293.1 NEVH01000250 PNF43885.1 KK852808 KDR16030.1 KQ971338 EFA02869.2 KQ982314 KYQ57956.1 BT127020 AEE61982.1 APGK01023288 APGK01023289 APGK01023290 KB740481 ENN80572.1 KB632431 ERL95517.1 GECZ01009507 JAS60262.1 KQ977754 KYN00081.1 GECU01027274 GECU01010869 JAS80432.1 JAS96837.1 KK107063 EZA61168.1 FX985318 BAX07331.1 GALA01001651 JAA93201.1 GALA01001666 JAA93186.1 ATLV01025637 KE525396 KFB52507.1 CH477692 EAT37202.1 Y11347 CH963857 EDW75792.2 GFDL01007624 JAV27421.1 HAAF01002611 CCP74437.1 AEYP01053153 JP017742 AES06340.1 DS232158 EDS36324.1 GAPW01001200 JAC12398.1 X58541 GAPW01001393 JAC12205.1 JXUM01026061 KQ560742 KXJ81047.1 CH902620 EDV31395.2 CH933807 EDW11093.1 GAPW01001199 JAC12399.1 JXUM01069686 JXUM01069687 JXUM01069688 JXUM01069689 KQ562564 KXJ75606.1 CM000362 CM002911 EDX07613.1 KMY94760.1 AEMK02000015 CH480816 EDW48403.1 AAKN02032075 OUUW01000004 SPP79442.1 FJ608558 ACO24971.1 CP012523 ALC38139.1 GEDC01008592 JAS28706.1 AQIB01127710 KE164168 EPQ15712.1 GGFK01000735 MBW34056.1 AP018254 BAZ06050.1 MTAW01000089 OUL32095.1 MTAX01000054 OUL26509.1 CM004470 OCT90552.1 GGFK01000742 MBW34063.1 RAZU01000172 RLQ66934.1 AGCU01051555 BC123188 AAI23189.1 CM000157 EDW88434.1 AC147986 CH466529 EDL19287.1 JH168609 EHB04710.1 GGFL01004633 MBW68811.1 CH916368 EDW03852.1 GGFM01006066 MBW26817.1 GGFJ01007696 MBW56837.1 AAPE02003478 GGFJ01005029 MBW54170.1 BC141762 AAI41763.1 CH479180 EDW29265.1 GGFK01007811 MBW41132.1 GGFM01006579 MBW27330.1 CH379060 EAL33942.2
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000192223
UP000078541
+ More
UP000235965 UP000027135 UP000007266 UP000075809 UP000019118 UP000030742 UP000078542 UP000053097 UP000030765 UP000008820 UP000008227 UP000007798 UP000000715 UP000002320 UP000075880 UP000248480 UP000069940 UP000249989 UP000007801 UP000007646 UP000009192 UP000233160 UP000000304 UP000248483 UP000192221 UP000001292 UP000005447 UP000268350 UP000092553 UP000079721 UP000029965 UP000221080 UP000218621 UP000195197 UP000195154 UP000186698 UP000273346 UP000189706 UP000007267 UP000233220 UP000002282 UP000000589 UP000006813 UP000001070 UP000001074 UP000069272 UP000008744 UP000001819
UP000235965 UP000027135 UP000007266 UP000075809 UP000019118 UP000030742 UP000078542 UP000053097 UP000030765 UP000008820 UP000008227 UP000007798 UP000000715 UP000002320 UP000075880 UP000248480 UP000069940 UP000249989 UP000007801 UP000007646 UP000009192 UP000233160 UP000000304 UP000248483 UP000192221 UP000001292 UP000005447 UP000268350 UP000092553 UP000079721 UP000029965 UP000221080 UP000218621 UP000195197 UP000195154 UP000186698 UP000273346 UP000189706 UP000007267 UP000233220 UP000002282 UP000000589 UP000006813 UP000001070 UP000001074 UP000069272 UP000008744 UP000001819
Interpro
Gene 3D
ProteinModelPortal
C0J8G2
A0A3G1VCH2
A0A2A4J5M2
A0A212F3H3
A0A194PVI0
A0A194R3B9
+ More
J7HLS7 S4NYK9 A0A2H1VQ07 X2J109 A0A1W4X1P5 A0A195F1F1 A0A2J7RSS5 A0A067R8V4 D2A2J9 A0A151XC37 J3JUV5 N6UPE2 U4UQY2 A0A1B6GCU3 A0A195CH67 A0A1B6JCM9 A0A026WZD2 A0A1V1FIT8 T1DGH8 T1DGG7 A0A084WQL3 Q16S06 P79335 B4MVS2 A0A1Q3FIJ5 U6CX60 G9KN61 A0A1S4JWS5 B0WX63 A0A023ESX4 A0A182J1Y2 P50449 A0A023ESG3 A0A2Y9E5Y9 A0A182H582 B3MJQ6 G3T7G7 B4KGA6 A0A023EV90 A0A182GK96 A0A2K6FA88 B4QBV3 I3LQQ6 A0A2Y9PNI9 A0A1W4V0X4 B4HNC1 H0UTC2 A0A3B0JGQ0 A0A286XDM0 C1JFL8 A0A0M3QT24 A0A1S2ZE36 A0A1B6DSP6 A0A0D9S037 S7Q4B4 A0A2M3ZZQ8 A0A2D0PLI2 A0A1Z4NK21 A0A252DWU7 A0A252DGG5 A0A1L8H381 A0A2M3ZZT6 A0A3L7HLU5 A0A1U7R3V0 K7FMQ9 A0A2K6TSH2 Q0IHE4 B4NWB7 G5E899 G5B5Z9 A0A2M4CUF6 B4JAP4 A0A2M3ZE19 A0A2M4BUU6 A0A1W4VWE9 G1NYA4 A0A182F4A5 A0A2M4BM55 A5D8P7 B4G7D2 A0A2M4AK75 A0A2M3ZFW7 Q29LV3
J7HLS7 S4NYK9 A0A2H1VQ07 X2J109 A0A1W4X1P5 A0A195F1F1 A0A2J7RSS5 A0A067R8V4 D2A2J9 A0A151XC37 J3JUV5 N6UPE2 U4UQY2 A0A1B6GCU3 A0A195CH67 A0A1B6JCM9 A0A026WZD2 A0A1V1FIT8 T1DGH8 T1DGG7 A0A084WQL3 Q16S06 P79335 B4MVS2 A0A1Q3FIJ5 U6CX60 G9KN61 A0A1S4JWS5 B0WX63 A0A023ESX4 A0A182J1Y2 P50449 A0A023ESG3 A0A2Y9E5Y9 A0A182H582 B3MJQ6 G3T7G7 B4KGA6 A0A023EV90 A0A182GK96 A0A2K6FA88 B4QBV3 I3LQQ6 A0A2Y9PNI9 A0A1W4V0X4 B4HNC1 H0UTC2 A0A3B0JGQ0 A0A286XDM0 C1JFL8 A0A0M3QT24 A0A1S2ZE36 A0A1B6DSP6 A0A0D9S037 S7Q4B4 A0A2M3ZZQ8 A0A2D0PLI2 A0A1Z4NK21 A0A252DWU7 A0A252DGG5 A0A1L8H381 A0A2M3ZZT6 A0A3L7HLU5 A0A1U7R3V0 K7FMQ9 A0A2K6TSH2 Q0IHE4 B4NWB7 G5E899 G5B5Z9 A0A2M4CUF6 B4JAP4 A0A2M3ZE19 A0A2M4BUU6 A0A1W4VWE9 G1NYA4 A0A182F4A5 A0A2M4BM55 A5D8P7 B4G7D2 A0A2M4AK75 A0A2M3ZFW7 Q29LV3
PDB
5CDZ
E-value=7.8097e-38,
Score=395
Ontologies
GO
GO:0005615
GO:0008233
GO:0010951
GO:0004867
GO:0090399
GO:0009611
GO:0035009
GO:0016747
GO:0045861
GO:0071222
GO:0050829
GO:0001525
GO:1901331
GO:2000352
GO:0045766
GO:0010757
GO:0048260
GO:0002020
GO:0062023
GO:2000098
GO:0070062
GO:0061044
GO:0035491
GO:0090026
GO:0014912
GO:0001300
GO:0097187
GO:0032757
GO:0005102
GO:1902042
GO:0033629
GO:0051918
GO:0010469
GO:0090136
GO:0005829
GO:0032940
GO:0061108
GO:0060384
GO:0031091
GO:0051966
GO:0060291
GO:0007596
GO:0045879
GO:0014067
GO:0042177
GO:0048711
GO:0008201
GO:0010766
GO:0021683
GO:0042628
GO:0090331
GO:0048505
GO:0031232
GO:0008285
GO:0031594
GO:0030308
GO:0050974
GO:0033363
PANTHER
Topology
Subcellular location
Secreted
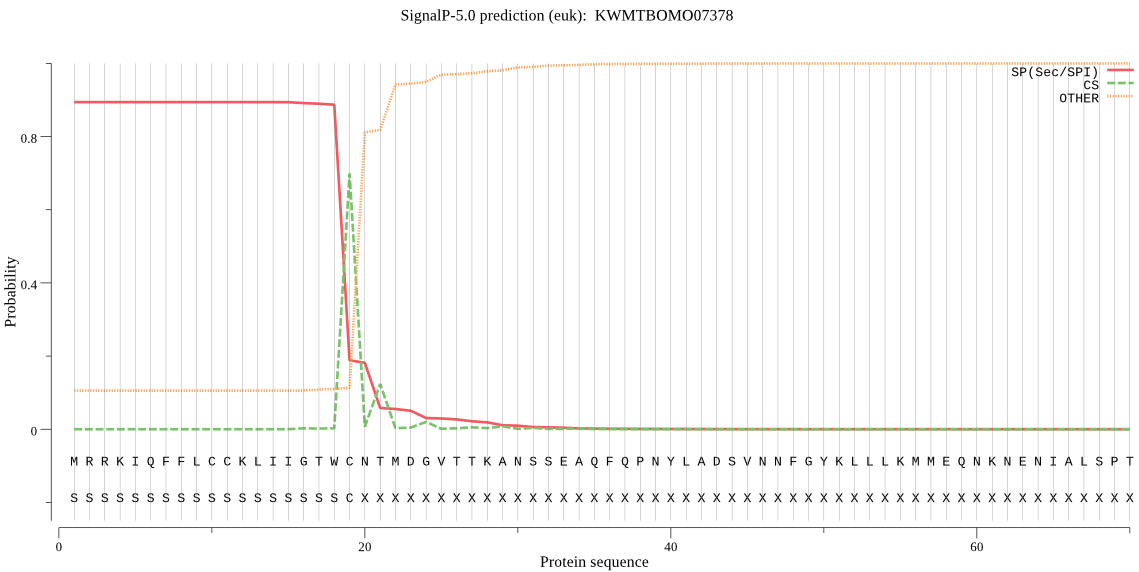
SignalP
Position: 1 - 19,
Likelihood: 0.893808
Length:
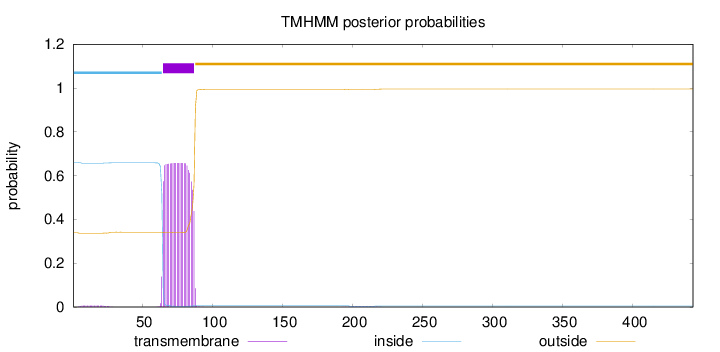
443
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
14.9506
Exp number, first 60 AAs:
0.13609
Total prob of N-in:
0.66108
inside
1 - 64
TMhelix
65 - 87
outside
88 - 443
Population Genetic Test Statistics
Pi
25.570577
Theta
21.930892
Tajima's D
0.774006
CLR
0.449003
CSRT
0.610069496525174
Interpretation
Uncertain
