Gene
KWMTBOMO07375
Pre Gene Modal
BGIBMGA005180
Annotation
PREDICTED:_DNA_repair_endonuclease_XPF_[Amyelois_transitella]
Full name
DNA repair endonuclease XPF
Alternative Name
Protein meiotic 9
Location in the cell
Nuclear Reliability : 3.474
Sequence
CDS
ATGTTGTCATTCGAAAAGCAAATCTTCATTGATGTGTTTGAAAAAGATGCTCTTGTAATAATGGCAAAAGGCCATAATTACAACAACATTGTGGCTAATTTGTTGTGGGTGCACAAGGATCCTGGTAATCTAGTTTTAGTTATAAACAGTAATGATAATGAAGAAAAATATTTTACTGAAAAAAATCAATTGACACCAGTACCCACACTTGGCAATGAAAGAGAGCGGTCGTATTTAGAGGGTGGAATTCATTTTGTATCAACTCGTATTTTGGTTGTGGATTTATTAAAAAACAGAGTGCCAATATCCCATATCACAGGCATTGTTGTTCTAAGAGCACACACAGTCTTAGAATCATGTCAGGAAGCATTTGCATTGAGGTTGTACAGGCAGAAAAATAAGACTGGATTTATTAAAGCATTTTCTAATTCACCTATCTCTTTCACGTTTGGATACCATCAAGTTGAGAAAGTTATGAGGGCGCTTTTTGTGACAGAACTATTTCTGTGGCCAAGGTTTCATGGCAACATTATTAGAAATTTAAAGAGTAGACAAGCTCAAGCTGAAGAACTTCACATTCCATTAACTACTAACATGAGCTACATTCAGACGTGTCTTCTTGACATTATGAACTACACTGTCAAACAATTGAAGAGCATTAACAGGTCTTTAGATATGAAAGAAATCACAACCGAGAATTGTATAACAAAGAAATTTCATAAGATCCTGCAATCGCAGCTGGATTGTGTGTGGCATCAACTTAGTACAAGAACAAAGGAATACATACAAGACCTAAAAATTCTCAGGAGCATGATTATTAACCTAATACACGATGACGCGGTGTCGTTATACGCGTTAGTCAGCAAGTACCGAACTCCTGAATACGCACAAATAAATTCTGGTTGGATCCTGCTCGAGTCTGCGGAGCAGCTGTTTAAATACGCCAAAGCACGTGTATTTAATAAAAAGAACGAATTCGATCCGGAGCCTTCACCGAAATGGAAGTACTTAAGCGATTTGTTATCCCAAGAAATACCAGACGAAGTAAACAAAAAGAAAGACAGTAACTTGAGCAACACTAAAATATTAATTCTCTGCCAAGACAATCGGACCTGTAGCCAGCTGAACCACTTCCTCACCATGGGGGCAAATAGATACCTGCTGTACACGGCTTTCAAACGTGAGATCCCCATAAATGCAGTAGCGACAAAATACGAAGACATAAAAAACACAGTTCCCGTAGAAACTAAAGATAAGACAGACAAAAAGTCGGATACAAAAGAAGGAAAATCTACCGATGAAGGCACACGTTTGGATGACTCGAAATATGAAGAATTGAACACAGAAGAATCGAGAACTAATTATCTGTTGACGCTAACACAAAGTTCAGCGCACTGCAGCAAAGATAATGCAACAGTTAATGAAAGTTTATTCGAACCCATTTCCCAGCTGGATAATTTAGATTTTACGAAAATCGAAGTAGAAACGCCCGTAATCTGCATCCAAACGTTCAAAGAAAACGGTAACAATCTCTCCTTGGAATACACTCTGGAAGCTTTGAGACCGGAATATGTTATACTGTATCACAGCGACGTGGCAGCTGTGCGCCAGCTGGAGATGTACGAGTGCAGAAAGAAACCGGAGGAGCCGATCTGCAAAGTGTACTTCCTGATACACGATCAAACAGTTGAGGAGCAAGCCTATCTGGCGGCATTGAAGAGAGAAAAACAAGCATTTGAATTGTTGATTCAGGCCAAAAGTGTCATGGTAGTTCCTTCATATCAAGACGGAAAAACAGATGAATACTTCCAATTAACAAACGAGGAAGAAGTGCAACCCACCGATTCAAGGAAAGCAGGTCAAAATCAAAGTAAAGAGAAACCGCTAGTGATAGTGGACATGAGAGAGTTTCGATCAGACTTACCGTCGCTGTTGCACAAGAGAGGCATTAATATCGAACCAGTTACGATAGCGATAGGTGATTACATACTGACCCCTGATATTTGCGTAGAAAGGAAATCTATATCCGATTTGATTGGATCGTTGAACTCTGGTCGCTTGTACACACAATGCACACAAATGTGCCGCAACTACAGTCGACCTATCCTGCTTATTGAATTTGATCAAAATAAACCTTTTAATCTTCAGGGTCATTTCGTGGTATCAACAGATATATCTGGAGCGGATATACAACAAAAACTACAACTTCTGACAATACATTTCCCTAGGCTGAAATTGGTATGGTCGCCAAGTCCGTATGCTACTGCTGAACTTTTCTATGAATTAAAGCAAGGCAGAAATAATCCCAATATTGCAGAAGCTATCGCTCTCAGTGGTGAAAATACCGCAGATGATATGAACTATGAGAGATACAATATATTGGTTCATGATTTCGTACAAAAACTACCTGGTGTTACATCAAAAAACATTAACAGAATTATGAACAAAGGTATCTCACTGGACCACTTGATAACTCTTACCAAAGAGGAATTATCTGATGTCGTAGAAAACAAAAATGAAGCCGAAACACTGTATTCAATACTTCACGTCAAAGCTCGTCCCATGGATGCCAAAGAAGAGAAATTCGGAAAGAAAAAGTTCACTGGAAAACTATATCGTAGTAAAAAATAA
Protein
MLSFEKQIFIDVFEKDALVIMAKGHNYNNIVANLLWVHKDPGNLVLVINSNDNEEKYFTEKNQLTPVPTLGNERERSYLEGGIHFVSTRILVVDLLKNRVPISHITGIVVLRAHTVLESCQEAFALRLYRQKNKTGFIKAFSNSPISFTFGYHQVEKVMRALFVTELFLWPRFHGNIIRNLKSRQAQAEELHIPLTTNMSYIQTCLLDIMNYTVKQLKSINRSLDMKEITTENCITKKFHKILQSQLDCVWHQLSTRTKEYIQDLKILRSMIINLIHDDAVSLYALVSKYRTPEYAQINSGWILLESAEQLFKYAKARVFNKKNEFDPEPSPKWKYLSDLLSQEIPDEVNKKKDSNLSNTKILILCQDNRTCSQLNHFLTMGANRYLLYTAFKREIPINAVATKYEDIKNTVPVETKDKTDKKSDTKEGKSTDEGTRLDDSKYEELNTEESRTNYLLTLTQSSAHCSKDNATVNESLFEPISQLDNLDFTKIEVETPVICIQTFKENGNNLSLEYTLEALRPEYVILYHSDVAAVRQLEMYECRKKPEEPICKVYFLIHDQTVEEQAYLAALKREKQAFELLIQAKSVMVVPSYQDGKTDEYFQLTNEEEVQPTDSRKAGQNQSKEKPLVIVDMREFRSDLPSLLHKRGINIEPVTIAIGDYILTPDICVERKSISDLIGSLNSGRLYTQCTQMCRNYSRPILLIEFDQNKPFNLQGHFVVSTDISGADIQQKLQLLTIHFPRLKLVWSPSPYATAELFYELKQGRNNPNIAEAIALSGENTADDMNYERYNILVHDFVQKLPGVTSKNINRIMNKGISLDHLITLTKEELSDVVENKNEAETLYSILHVKARPMDAKEEKFGKKKFTGKLYRSKK
Summary
Subunit
Heterodimer (By similarity). Interacts with hdm.
Similarity
Belongs to the XPF family.
Keywords
Complete proteome
DNA damage
DNA repair
DNA-binding
Endonuclease
Hydrolase
Meiosis
Nuclease
Nucleus
Reference proteome
Feature
chain DNA repair endonuclease XPF
Uniprot
A0A3S2NRS2
A0A212F3J3
A0A194PTM5
A0A194R2V2
A0A2H1VNE8
A0A1E1WJ72
+ More
A0A2A4IUY9 A0A1I8P6G3 W8BG68 A0A1I8M666 Q29FQ4 A0A1W4V1G6 A0A1A9ZVD3 A0A0A1WM55 B4GTY4 A0A1Y1JT47 A0A034WDQ6 A0A0K8UYR4 A0A0M3QZ63 A0A1A9XAJ4 A0A1B0ATA9 T2FFI3 Q24087 A0A1A9UPJ2 A0A1A9WG73 A0A3B0K781 B4L6Q4 B4Q069 B4NP89 B3NU19 A0A336M4G7 B3MSI5 B4JMQ3 A0A0L0CGH6 B4M7P7 A0A336MFC1 Q17J54 A0A1W4WJ37 A0A1J1IGF4 D6WJT6 A0A0J7KEA4 A0A232F607 U4TZA3 N6TEU9 A0A1B6EY17 A0A026WM20 E2B2N4 A0A3L8E4Q4 E2A3B3 A0A0N0BE83 A0A2Z5U214 A0A067R0V8 A0A2A3EF28 A0A1B0FGJ6 A0A154PG49 A0A1B6E0K5 A0A2S2QUC3 A0A2H8TG83 A0A224XJV8 A0A088AAW9 A0A0A9ZJ13 J9JL14 A0A158NY48 T1HKQ6 A0A1Y1JT32 A0A151IE27 V4C7S3 A0A1S3J1P2 A0A1S3IMB0 K1P9V1 A0A0P4W933 A0A210R1E3 A0A2C9KG40 A0A1Z5LEM3 A0A2C9KG37 A0A2T7NQ56 H3ABZ1 R7TEW3 A0A131XJX8 L7M9M1 A0A224YUW8 A0A131YWV5 A0A2Y9DFR3 L7MLT1 A0A182G9Y0 F6R8S0 A0A0K2T0T9 A0A3P8WV94 A0A3Q0RCE3 A0A3B5AHN8 A0A3Q3BI53 A0A0L8HK36 A0A3B4C2C1 A0A3Q1I007 U3J3D1 A0A3Q2ZFV8 A0A3Q3GJQ8 A0A3Q3VSZ3 A0A2K6ES99 A0A2I4D8D4
A0A2A4IUY9 A0A1I8P6G3 W8BG68 A0A1I8M666 Q29FQ4 A0A1W4V1G6 A0A1A9ZVD3 A0A0A1WM55 B4GTY4 A0A1Y1JT47 A0A034WDQ6 A0A0K8UYR4 A0A0M3QZ63 A0A1A9XAJ4 A0A1B0ATA9 T2FFI3 Q24087 A0A1A9UPJ2 A0A1A9WG73 A0A3B0K781 B4L6Q4 B4Q069 B4NP89 B3NU19 A0A336M4G7 B3MSI5 B4JMQ3 A0A0L0CGH6 B4M7P7 A0A336MFC1 Q17J54 A0A1W4WJ37 A0A1J1IGF4 D6WJT6 A0A0J7KEA4 A0A232F607 U4TZA3 N6TEU9 A0A1B6EY17 A0A026WM20 E2B2N4 A0A3L8E4Q4 E2A3B3 A0A0N0BE83 A0A2Z5U214 A0A067R0V8 A0A2A3EF28 A0A1B0FGJ6 A0A154PG49 A0A1B6E0K5 A0A2S2QUC3 A0A2H8TG83 A0A224XJV8 A0A088AAW9 A0A0A9ZJ13 J9JL14 A0A158NY48 T1HKQ6 A0A1Y1JT32 A0A151IE27 V4C7S3 A0A1S3J1P2 A0A1S3IMB0 K1P9V1 A0A0P4W933 A0A210R1E3 A0A2C9KG40 A0A1Z5LEM3 A0A2C9KG37 A0A2T7NQ56 H3ABZ1 R7TEW3 A0A131XJX8 L7M9M1 A0A224YUW8 A0A131YWV5 A0A2Y9DFR3 L7MLT1 A0A182G9Y0 F6R8S0 A0A0K2T0T9 A0A3P8WV94 A0A3Q0RCE3 A0A3B5AHN8 A0A3Q3BI53 A0A0L8HK36 A0A3B4C2C1 A0A3Q1I007 U3J3D1 A0A3Q2ZFV8 A0A3Q3GJQ8 A0A3Q3VSZ3 A0A2K6ES99 A0A2I4D8D4
EC Number
3.1.-.-
Pubmed
22118469
26354079
24495485
25315136
15632085
17994087
+ More
25830018 28004739 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 8647398 12537569 18957705 17550304 26108605 17510324 18362917 19820115 28648823 23537049 24508170 20798317 30249741 26760975 24845553 25401762 21347285 23254933 22992520 28812685 15562597 28528879 9215903 28049606 25576852 28797301 26830274 26483478 19892987 24487278 23749191
25830018 28004739 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 8647398 12537569 18957705 17550304 26108605 17510324 18362917 19820115 28648823 23537049 24508170 20798317 30249741 26760975 24845553 25401762 21347285 23254933 22992520 28812685 15562597 28528879 9215903 28049606 25576852 28797301 26830274 26483478 19892987 24487278 23749191
EMBL
RSAL01000116
RVE46864.1
AGBW02010564
OWR48283.1
KQ459592
KPI96766.1
+ More
KQ460845 KPJ12027.1 ODYU01003476 SOQ42317.1 GDQN01004143 JAT86911.1 NWSH01006888 PCG63198.1 GAMC01006260 JAC00296.1 CH379065 EAL31525.2 GBXI01014809 JAC99482.1 CH479190 EDW26004.1 GEZM01101618 JAV52442.1 GAKP01006133 JAC52819.1 GDHF01020616 JAI31698.1 CP012528 ALC48873.1 JXJN01003231 BT150257 AE014298 AGV77159.1 AHN59330.1 U27181 BT001319 OUUW01000018 SPP89163.1 CH933812 EDW06050.1 CM000162 EDX01221.1 CH964291 EDW86329.1 CH954180 EDV45795.1 UFQT01000387 SSX23823.1 CH902622 EDV34740.2 CH916371 EDV91996.1 JRES01000430 KNC31345.1 CH940653 EDW62814.1 UFQT01001121 SSX29005.1 CH477234 EAT46753.1 CVRI01000048 CRK98620.1 KQ971343 EFA04567.1 LBMM01008502 KMQ88823.1 NNAY01000882 OXU26075.1 KB631899 ERL86939.1 APGK01040820 KB740984 ENN76293.1 GECZ01027038 GECZ01020036 JAS42731.1 JAS49733.1 KK107168 EZA56134.1 GL445190 EFN90056.1 QOIP01000001 RLU27522.1 GL436354 EFN72085.1 KQ435844 KOX71272.1 FX985834 BBA93721.1 KK852822 KDR15512.1 KZ288267 PBC30084.1 CCAG010010420 KQ434886 KZC10170.1 GEDC01005832 JAS31466.1 GGMS01011529 MBY80732.1 GFXV01001245 MBW13050.1 GFTR01008115 JAW08311.1 GBHO01000389 GBRD01010326 JAG43215.1 JAG55498.1 ABLF02040901 ADTU01003143 ADTU01003144 ACPB03014634 GEZM01101617 JAV52444.1 KQ977914 KYM98817.1 KB201304 ESO97764.1 JH815775 EKC18198.1 GDRN01071374 GDRN01071373 JAI63699.1 NEDP02000888 OWF54777.1 GFJQ02001111 JAW05859.1 PZQS01000010 PVD23305.1 AFYH01147897 AFYH01147898 AFYH01147899 AMQN01003141 KB311062 ELT90017.1 GEFH01002740 JAP65841.1 GACK01005230 JAA59804.1 GFPF01007255 MAA18401.1 GEDV01006156 JAP82401.1 GACK01000069 JAA64965.1 JXUM01049950 JXUM01049951 JXUM01049952 JXUM01049953 JXUM01049954 JXUM01049955 JXUM01049956 JXUM01049957 KQ561628 KXJ77953.1 HACA01002277 CDW19638.1 KQ417942 KOF89596.1 ADON01130592 ADON01130593 ADON01130594
KQ460845 KPJ12027.1 ODYU01003476 SOQ42317.1 GDQN01004143 JAT86911.1 NWSH01006888 PCG63198.1 GAMC01006260 JAC00296.1 CH379065 EAL31525.2 GBXI01014809 JAC99482.1 CH479190 EDW26004.1 GEZM01101618 JAV52442.1 GAKP01006133 JAC52819.1 GDHF01020616 JAI31698.1 CP012528 ALC48873.1 JXJN01003231 BT150257 AE014298 AGV77159.1 AHN59330.1 U27181 BT001319 OUUW01000018 SPP89163.1 CH933812 EDW06050.1 CM000162 EDX01221.1 CH964291 EDW86329.1 CH954180 EDV45795.1 UFQT01000387 SSX23823.1 CH902622 EDV34740.2 CH916371 EDV91996.1 JRES01000430 KNC31345.1 CH940653 EDW62814.1 UFQT01001121 SSX29005.1 CH477234 EAT46753.1 CVRI01000048 CRK98620.1 KQ971343 EFA04567.1 LBMM01008502 KMQ88823.1 NNAY01000882 OXU26075.1 KB631899 ERL86939.1 APGK01040820 KB740984 ENN76293.1 GECZ01027038 GECZ01020036 JAS42731.1 JAS49733.1 KK107168 EZA56134.1 GL445190 EFN90056.1 QOIP01000001 RLU27522.1 GL436354 EFN72085.1 KQ435844 KOX71272.1 FX985834 BBA93721.1 KK852822 KDR15512.1 KZ288267 PBC30084.1 CCAG010010420 KQ434886 KZC10170.1 GEDC01005832 JAS31466.1 GGMS01011529 MBY80732.1 GFXV01001245 MBW13050.1 GFTR01008115 JAW08311.1 GBHO01000389 GBRD01010326 JAG43215.1 JAG55498.1 ABLF02040901 ADTU01003143 ADTU01003144 ACPB03014634 GEZM01101617 JAV52444.1 KQ977914 KYM98817.1 KB201304 ESO97764.1 JH815775 EKC18198.1 GDRN01071374 GDRN01071373 JAI63699.1 NEDP02000888 OWF54777.1 GFJQ02001111 JAW05859.1 PZQS01000010 PVD23305.1 AFYH01147897 AFYH01147898 AFYH01147899 AMQN01003141 KB311062 ELT90017.1 GEFH01002740 JAP65841.1 GACK01005230 JAA59804.1 GFPF01007255 MAA18401.1 GEDV01006156 JAP82401.1 GACK01000069 JAA64965.1 JXUM01049950 JXUM01049951 JXUM01049952 JXUM01049953 JXUM01049954 JXUM01049955 JXUM01049956 JXUM01049957 KQ561628 KXJ77953.1 HACA01002277 CDW19638.1 KQ417942 KOF89596.1 ADON01130592 ADON01130593 ADON01130594
Proteomes
UP000283053
UP000007151
UP000053268
UP000053240
UP000218220
UP000095300
+ More
UP000095301 UP000001819 UP000192221 UP000092445 UP000008744 UP000092553 UP000092443 UP000092460 UP000000803 UP000078200 UP000091820 UP000268350 UP000009192 UP000002282 UP000007798 UP000008711 UP000007801 UP000001070 UP000037069 UP000008792 UP000008820 UP000192223 UP000183832 UP000007266 UP000036403 UP000215335 UP000030742 UP000019118 UP000053097 UP000008237 UP000279307 UP000000311 UP000053105 UP000027135 UP000242457 UP000092444 UP000076502 UP000005203 UP000007819 UP000005205 UP000015103 UP000078542 UP000030746 UP000085678 UP000005408 UP000242188 UP000076420 UP000245119 UP000008672 UP000014760 UP000248480 UP000069940 UP000249989 UP000002281 UP000265120 UP000261340 UP000261400 UP000264800 UP000053454 UP000261440 UP000265040 UP000016666 UP000264820 UP000261660 UP000261620 UP000233160 UP000192220
UP000095301 UP000001819 UP000192221 UP000092445 UP000008744 UP000092553 UP000092443 UP000092460 UP000000803 UP000078200 UP000091820 UP000268350 UP000009192 UP000002282 UP000007798 UP000008711 UP000007801 UP000001070 UP000037069 UP000008792 UP000008820 UP000192223 UP000183832 UP000007266 UP000036403 UP000215335 UP000030742 UP000019118 UP000053097 UP000008237 UP000279307 UP000000311 UP000053105 UP000027135 UP000242457 UP000092444 UP000076502 UP000005203 UP000007819 UP000005205 UP000015103 UP000078542 UP000030746 UP000085678 UP000005408 UP000242188 UP000076420 UP000245119 UP000008672 UP000014760 UP000248480 UP000069940 UP000249989 UP000002281 UP000265120 UP000261340 UP000261400 UP000264800 UP000053454 UP000261440 UP000265040 UP000016666 UP000264820 UP000261660 UP000261620 UP000233160 UP000192220
Interpro
Gene 3D
ProteinModelPortal
A0A3S2NRS2
A0A212F3J3
A0A194PTM5
A0A194R2V2
A0A2H1VNE8
A0A1E1WJ72
+ More
A0A2A4IUY9 A0A1I8P6G3 W8BG68 A0A1I8M666 Q29FQ4 A0A1W4V1G6 A0A1A9ZVD3 A0A0A1WM55 B4GTY4 A0A1Y1JT47 A0A034WDQ6 A0A0K8UYR4 A0A0M3QZ63 A0A1A9XAJ4 A0A1B0ATA9 T2FFI3 Q24087 A0A1A9UPJ2 A0A1A9WG73 A0A3B0K781 B4L6Q4 B4Q069 B4NP89 B3NU19 A0A336M4G7 B3MSI5 B4JMQ3 A0A0L0CGH6 B4M7P7 A0A336MFC1 Q17J54 A0A1W4WJ37 A0A1J1IGF4 D6WJT6 A0A0J7KEA4 A0A232F607 U4TZA3 N6TEU9 A0A1B6EY17 A0A026WM20 E2B2N4 A0A3L8E4Q4 E2A3B3 A0A0N0BE83 A0A2Z5U214 A0A067R0V8 A0A2A3EF28 A0A1B0FGJ6 A0A154PG49 A0A1B6E0K5 A0A2S2QUC3 A0A2H8TG83 A0A224XJV8 A0A088AAW9 A0A0A9ZJ13 J9JL14 A0A158NY48 T1HKQ6 A0A1Y1JT32 A0A151IE27 V4C7S3 A0A1S3J1P2 A0A1S3IMB0 K1P9V1 A0A0P4W933 A0A210R1E3 A0A2C9KG40 A0A1Z5LEM3 A0A2C9KG37 A0A2T7NQ56 H3ABZ1 R7TEW3 A0A131XJX8 L7M9M1 A0A224YUW8 A0A131YWV5 A0A2Y9DFR3 L7MLT1 A0A182G9Y0 F6R8S0 A0A0K2T0T9 A0A3P8WV94 A0A3Q0RCE3 A0A3B5AHN8 A0A3Q3BI53 A0A0L8HK36 A0A3B4C2C1 A0A3Q1I007 U3J3D1 A0A3Q2ZFV8 A0A3Q3GJQ8 A0A3Q3VSZ3 A0A2K6ES99 A0A2I4D8D4
A0A2A4IUY9 A0A1I8P6G3 W8BG68 A0A1I8M666 Q29FQ4 A0A1W4V1G6 A0A1A9ZVD3 A0A0A1WM55 B4GTY4 A0A1Y1JT47 A0A034WDQ6 A0A0K8UYR4 A0A0M3QZ63 A0A1A9XAJ4 A0A1B0ATA9 T2FFI3 Q24087 A0A1A9UPJ2 A0A1A9WG73 A0A3B0K781 B4L6Q4 B4Q069 B4NP89 B3NU19 A0A336M4G7 B3MSI5 B4JMQ3 A0A0L0CGH6 B4M7P7 A0A336MFC1 Q17J54 A0A1W4WJ37 A0A1J1IGF4 D6WJT6 A0A0J7KEA4 A0A232F607 U4TZA3 N6TEU9 A0A1B6EY17 A0A026WM20 E2B2N4 A0A3L8E4Q4 E2A3B3 A0A0N0BE83 A0A2Z5U214 A0A067R0V8 A0A2A3EF28 A0A1B0FGJ6 A0A154PG49 A0A1B6E0K5 A0A2S2QUC3 A0A2H8TG83 A0A224XJV8 A0A088AAW9 A0A0A9ZJ13 J9JL14 A0A158NY48 T1HKQ6 A0A1Y1JT32 A0A151IE27 V4C7S3 A0A1S3J1P2 A0A1S3IMB0 K1P9V1 A0A0P4W933 A0A210R1E3 A0A2C9KG40 A0A1Z5LEM3 A0A2C9KG37 A0A2T7NQ56 H3ABZ1 R7TEW3 A0A131XJX8 L7M9M1 A0A224YUW8 A0A131YWV5 A0A2Y9DFR3 L7MLT1 A0A182G9Y0 F6R8S0 A0A0K2T0T9 A0A3P8WV94 A0A3Q0RCE3 A0A3B5AHN8 A0A3Q3BI53 A0A0L8HK36 A0A3B4C2C1 A0A3Q1I007 U3J3D1 A0A3Q2ZFV8 A0A3Q3GJQ8 A0A3Q3VSZ3 A0A2K6ES99 A0A2I4D8D4
PDB
2BGW
E-value=2.2671e-12,
Score=178
Ontologies
PATHWAY
GO
GO:0006281
GO:0004520
GO:0003697
GO:0004519
GO:0003677
GO:0004518
GO:0032991
GO:0006298
GO:0000712
GO:0006302
GO:0046982
GO:0016321
GO:0030716
GO:0006289
GO:0006974
GO:0045132
GO:0006310
GO:0007131
GO:0005634
GO:0007143
GO:0000724
GO:0000014
GO:0006296
GO:0003684
GO:1901255
GO:0000110
GO:0016021
GO:0006303
GO:0000784
GO:1905768
GO:0047485
GO:0005669
GO:0061819
GO:0034644
GO:0006295
GO:0070522
GO:0042802
GO:0001094
GO:0008022
GO:1990599
GO:0009650
GO:1905765
GO:0010506
GO:1904357
GO:0006259
GO:0043022
GO:0005488
GO:0003707
GO:0051537
Topology
Subcellular location
Nucleus
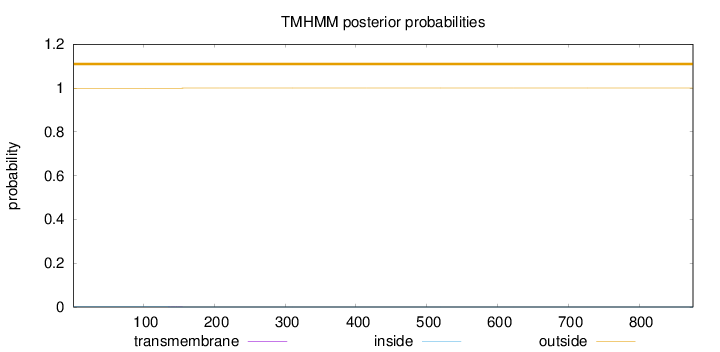
Length:
876
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02191
Exp number, first 60 AAs:
0.00054
Total prob of N-in:
0.00112
outside
1 - 876
Population Genetic Test Statistics
Pi
23.319878
Theta
21.220978
Tajima's D
0.432062
CLR
0.508044
CSRT
0.498325083745813
Interpretation
Uncertain
