Gene
KWMTBOMO07371
Pre Gene Modal
BGIBMGA010354
Annotation
hypothetical_protein_KGM_15726_[Danaus_plexippus]
Full name
Protein turtle
Location in the cell
Extracellular Reliability : 1.41 PlasmaMembrane Reliability : 1.028
Sequence
CDS
ATGGGCTGGCGCGCCGTGCAACCGCCGCACATCGCCGCCGGCTTGCTCCTCCTGCTGCTTGTCAATCTACCAGTGACCTGCCATCAAGACCACCAAGATGCCGTGCACATCACGGCTATCCTCGGTGAAAGTGTCGTGTTCAACTGTCAGGTGGATTTTCCCGAAGATATCCCGGTGCCGTACGTGTTGCAGTGGGAGAAGAAGGTAGGCGAAACGGGACAGGACATCCCGATCTACATCTGGTACGAGAGCTATCCGACGCATAGCGGGGAGGGCTACGAAGGCAGAGTGTCCCGCGTCGCAACCGACTCACCGTACGGCGCCGCAAGCCTCAACCTCACCAACATTCGAGAATCGGATCAGGGTTGGTACGAGTGTAAAGTGGTTTTTCTCAACCGATCACCAAACCAGCATAAGAATGGTACATGGTTCCACTTGGACGTGCACGCGCCACCTCGCTTCTCCATCACACCGGAAGACATTATATACGTTAATTTAGGCGATGCGATTATACTTAACTGCCAAGCAGAGGGAACACCAACTCCTGAAATACTATGGTATAAAGATGCAAACCCCGTCGAACCATCCGGTACTGTAGGTATATTTAACGATGGAACCGAACTGCGAATTAGTAACATTCGACACGAAGACATCGGTGATTACACGTGTATAGCGCGGAATGGAGAAGGTCAGGTATCTCACACGGCACGTGTCATTATCGCTGGTGGAGCTGTCATAACCATGCCACCGACAAATCAAACTAAACTGGAAGGAGAGAAAGTCCAGTTTTCGTGTGAAGCGAAGGCTCTGCCCGGAAATGTGACTGTGAAGTGGTTTCGTGAGGGTGCACCTGTGGCCGAGGTGGCGGCTTTGGAAACAAGAGTCACCATCCGCAGAGACGGAGCTCTTGTCATTAACCCTGTAGCTGCTGACGACTCCGGACAGTATTTGTGTGAAGTTTCTAATGGCATCGGAGACCCACAAAGCGCATCGGCCTATCTTAATGTTGAATATCCGGCGAAAGTAACCTTTACACCCACAGTTCAGTATTTGCCCTTCCGTTTGGCCGGAGTGGTTCAATGTTACATAAAAGCCAATCCACCTCTGCAGTACGTTACTTGGACAAAAGACAAGAGATTATTGGAGCCTTATCAAACGAAGGACATTGTTATAATGAACAACGGATCATTGTTATTTACCAGAGTTAACCAAAATCACCAGGGCCGGTACACTTGCACTCCTTATAACGCTCAAGGAACCCAAGGTTCTTCTGGTCCAATGGAAGTTCTAGTTCGGAAACCACCGGTGTTCACAGTGGAACCCGAACCATTATATCAGAGGAAGGTCGGTGAATCTGTAGAGATGCACTGCGAAGCACAAGAGTCTGAAGGCACACAGCGACCGAGCGTGACGTGGCGGCGGCGTGACGGTCTACCGCTTCAGAAGAACAGAGTCCGAGCTGTGGGCGGAAACATCACCATCGATACACTGCGCCGACAAGACTTTGGTATCTACCAATGTGTCGCGTCCAATGAGGTGGCAACAATCGTAGCGGACACGCAATTAGTGATAGAAGGCACGCAACCCCATGCTCCGTACAATGTGTCTGGAACAGCAACTGAGTTCCAAGTTACTTTGCGGTGGCAACCAGGATACGCCGGAGGTCCTGACTACAAACAGGACTATACGATATGGTACAGGGAAGCTGGATTTTCTGAGTGGACCAAAGTTCCCGTGACGCCCCCTGGTGCTACATCTGTAACAATTAACCGTCTACAACCTGGGACAACATACGAGTTTCAAGTAAATAGCAAGAATACTATAGGCGAAGGAATGATGAGTAAAGCAATTACAATAAGAACGCTCGATGTAGGTGCCAAGCCAAAGGTACCCACGACCGCCGCTGGTCCTATCGACGAAAAGATATTTCAAAACGCACCTGAAGGCTCCGGCCCGAAACCAGGTCCCCCACGGAATCTGACAGTGACAGAGGTTCATAATGGATTCTTAATAACTTGGCAAGGACCTTTAGAACGTGCGCAGCTTGTTCAGTACTACACCATCAAGTACCGCACTGACGCGCAGTGGAAGACCCTCAACAGGGGCCAGATACGACCTGAAGAGACCAGTTATTTAGTTAAGAACTTAGTCGGCGGGCGTACTTATTATTTTCGAGTACTCGCGAACTCGGCGACCAGCTACGAGAGTTCGGACGAAGTGAGGTTCCCGGTGCCGGCACGCGTGAAGCACAAAGCGATCACGGCTGGTGTCGTGGGTGGGATTCTGTTCTTCATCGTCGCCATCATACTGTCCGTGTGTGCCGTCAAGATATGTAACAAGCGTAAACGGCGTAAGCAGGAGAAAGAAATTATACGATTACAGCATACAACATGGTAG
Protein
MGWRAVQPPHIAAGLLLLLLVNLPVTCHQDHQDAVHITAILGESVVFNCQVDFPEDIPVPYVLQWEKKVGETGQDIPIYIWYESYPTHSGEGYEGRVSRVATDSPYGAASLNLTNIRESDQGWYECKVVFLNRSPNQHKNGTWFHLDVHAPPRFSITPEDIIYVNLGDAIILNCQAEGTPTPEILWYKDANPVEPSGTVGIFNDGTELRISNIRHEDIGDYTCIARNGEGQVSHTARVIIAGGAVITMPPTNQTKLEGEKVQFSCEAKALPGNVTVKWFREGAPVAEVAALETRVTIRRDGALVINPVAADDSGQYLCEVSNGIGDPQSASAYLNVEYPAKVTFTPTVQYLPFRLAGVVQCYIKANPPLQYVTWTKDKRLLEPYQTKDIVIMNNGSLLFTRVNQNHQGRYTCTPYNAQGTQGSSGPMEVLVRKPPVFTVEPEPLYQRKVGESVEMHCEAQESEGTQRPSVTWRRRDGLPLQKNRVRAVGGNITIDTLRRQDFGIYQCVASNEVATIVADTQLVIEGTQPHAPYNVSGTATEFQVTLRWQPGYAGGPDYKQDYTIWYREAGFSEWTKVPVTPPGATSVTINRLQPGTTYEFQVNSKNTIGEGMMSKAITIRTLDVGAKPKVPTTAAGPIDEKIFQNAPEGSGPKPGPPRNLTVTEVHNGFLITWQGPLERAQLVQYYTIKYRTDAQWKTLNRGQIRPEETSYLVKNLVGGRTYYFRVLANSATSYESSDEVRFPVPARVKHKAITAGVVGGILFFIVAIILSVCAVKICNKRKRRKQEKEIIRLQHTTW
Summary
Description
Essential protein that plays a role in the establishment of coordinated motor control (PubMed:11312296). In the developing eye, involved in axonal targeting of the R7 photoreceptor (PubMed:24174674).
Subunit
Interacts with bdl.
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Belongs to the immunoglobulin superfamily. Turtle family.
Belongs to the immunoglobulin superfamily. Turtle family.
Keywords
Alternative splicing
Complete proteome
Disulfide bond
Immunoglobulin domain
Membrane
Neurogenesis
Reference proteome
Repeat
Transmembrane
Transmembrane helix
Feature
chain Protein turtle
splice variant In isoform 3.
splice variant In isoform 3.
Uniprot
A0A1U9XRB3
A0A2A4J3J1
A0A212F3G4
A0A3S2NX96
A0A2A4J2M0
A0A194PUV0
+ More
A0A194R837 A0A2H1V9G2 A0A2A4J4G3 Q7Q3K8 A0A1S4J7Q9 A0A1S4GWQ1 A0A1S4FDQ5 A0A084VP30 Q0IF39 A0A182PJ73 A0A182JK20 A0A182F4L4 A0A182HV77 A0A1Y1MQA9 A0A182NE36 A0A1Y1MQ97 A0A1J1ID06 A0A182UV49 A0A182RBL8 A0A1Y1MTP8 A0A1Y1MTN2 A0A182WT84 A0A182YGU3 A0A154PTC6 A0A1Y1MQ85 A0A1Y1MRZ4 A0A182M3Y9 A0A336LVW2 W8B3X5 A0A1B6M0G0 A0A0Q9WKY0 A0A0Q9WL38 A0A182G7G0 A0A336M4C3 B4MDD0 A0A034VBV9 W8BDV1 V9I653 B4N0N6 A0A0R1DK60 A0A0J9QVH7 A0A0R3NRK4 Q967D7-5 W8AHL1 A0A139WJV7 A0A0M3QT07 A0A0R1DJ34 B4I374 Q967D7-2 E1JHS9 Q967D7-4 A0A0J9QWH3 A0A0R3NY41 A0A0R3NRT6 A0A139WJK5 A0A0R1DJ77 A0A3B0JZJ0 Q967D7 E1JHT0 A0A1B6CWC2 A0A1W4UQ05 A0A1W4V3Y0 A0A0A1XG79 A0A0P9A3B3 B4JD36 A0A1B6DNG5 A0A1W4UQJ1 A0A0A1X297 A0A0R1DQD2 A0A0Q5W9B6 A0A1W4UQI6 A0A1W4V2S8 W5JFI5 A0A1W4V2G1 A0A0J9TF92 A0A0R3NXQ3 T1HFR6 A0A0Q5W961 A0A0P8XE88 A0A1B6E919 A0A1B6C0M6 A0A0A1X554 A0A0R1DJD6 A0A0Q5WKV0 A0A0Q5WID7 A0A0C9R6F7 A0A0C9S214 B4NYI7 A0A0R1DQN2 A0A0R3NRN3 A0A0J9QVG8 A0A0R3NWW3 M9PCI8 E0VN01
A0A194R837 A0A2H1V9G2 A0A2A4J4G3 Q7Q3K8 A0A1S4J7Q9 A0A1S4GWQ1 A0A1S4FDQ5 A0A084VP30 Q0IF39 A0A182PJ73 A0A182JK20 A0A182F4L4 A0A182HV77 A0A1Y1MQA9 A0A182NE36 A0A1Y1MQ97 A0A1J1ID06 A0A182UV49 A0A182RBL8 A0A1Y1MTP8 A0A1Y1MTN2 A0A182WT84 A0A182YGU3 A0A154PTC6 A0A1Y1MQ85 A0A1Y1MRZ4 A0A182M3Y9 A0A336LVW2 W8B3X5 A0A1B6M0G0 A0A0Q9WKY0 A0A0Q9WL38 A0A182G7G0 A0A336M4C3 B4MDD0 A0A034VBV9 W8BDV1 V9I653 B4N0N6 A0A0R1DK60 A0A0J9QVH7 A0A0R3NRK4 Q967D7-5 W8AHL1 A0A139WJV7 A0A0M3QT07 A0A0R1DJ34 B4I374 Q967D7-2 E1JHS9 Q967D7-4 A0A0J9QWH3 A0A0R3NY41 A0A0R3NRT6 A0A139WJK5 A0A0R1DJ77 A0A3B0JZJ0 Q967D7 E1JHT0 A0A1B6CWC2 A0A1W4UQ05 A0A1W4V3Y0 A0A0A1XG79 A0A0P9A3B3 B4JD36 A0A1B6DNG5 A0A1W4UQJ1 A0A0A1X297 A0A0R1DQD2 A0A0Q5W9B6 A0A1W4UQI6 A0A1W4V2S8 W5JFI5 A0A1W4V2G1 A0A0J9TF92 A0A0R3NXQ3 T1HFR6 A0A0Q5W961 A0A0P8XE88 A0A1B6E919 A0A1B6C0M6 A0A0A1X554 A0A0R1DJD6 A0A0Q5WKV0 A0A0Q5WID7 A0A0C9R6F7 A0A0C9S214 B4NYI7 A0A0R1DQN2 A0A0R3NRN3 A0A0J9QVG8 A0A0R3NWW3 M9PCI8 E0VN01
Pubmed
EMBL
KY350198
AQZ26950.1
NWSH01003437
PCG66306.1
AGBW02010564
OWR48280.1
+ More
RSAL01000116 RVE46867.1 PCG66305.1 KQ459592 KPI96763.1 KQ460845 KPJ12031.1 ODYU01001360 SOQ37426.1 PCG66303.1 AAAB01008964 EAA12846.4 ATLV01014971 ATLV01014972 KE524999 KFB39724.1 CH477394 EAT41900.1 APCN01002500 GEZM01024234 JAV87842.1 GEZM01024223 JAV87859.1 CVRI01000044 CRK96838.1 GEZM01024232 JAV87845.1 GEZM01024228 JAV87850.1 KQ435102 KZC14664.1 GEZM01024229 JAV87849.1 GEZM01024230 JAV87848.1 AXCM01008531 UFQT01000144 SSX20849.1 GAMC01018609 JAB87946.1 GEBQ01010558 JAT29419.1 CH940661 KRF85516.1 KRF85517.1 JXUM01008362 JXUM01008363 JXUM01008364 KQ560259 KXJ83503.1 SSX20848.1 EDW71191.2 GAKP01019355 JAC39597.1 GAMC01018611 JAB87944.1 JR035185 AEY56260.1 CH963920 EDW77649.2 CM000157 KRJ97262.1 CM002910 KMY88043.1 CH379059 KRT03730.1 AJ312132 AJ312133 AJ312134 AJ312135 AE014134 BT010247 GAMC01018610 JAB87945.1 KQ971338 KYB28097.1 CP012523 ALC38083.1 KRJ97260.1 CH480820 EDW54219.1 ACZ94160.1 KMY88039.1 KRT03731.1 KRT03722.1 KYB28096.1 KRJ97258.1 OUUW01000004 SPP79099.1 ACZ94159.1 AGB92556.1 GEDC01019570 JAS17728.1 GBXI01004372 JAD09920.1 CH902620 KPU73026.1 CH916368 EDW04280.1 GEDC01010089 JAS27209.1 GBXI01008828 JAD05464.1 KRJ97264.1 CH954177 KQS70076.1 ADMH02001548 ETN62103.1 KMY88040.1 KRT03728.1 ACPB03018591 KQS70081.1 KPU73025.1 GEDC01002875 JAS34423.1 GEDC01030374 GEDC01023640 GEDC01014706 JAS06924.1 JAS13658.1 JAS22592.1 GBXI01008427 JAD05865.1 KRJ97256.1 KQS70082.1 KQS70078.1 GBYB01003570 JAG73337.1 GBYB01014906 JAG84673.1 EDW87620.1 KRJ97254.1 KRJ97255.1 KRJ97259.1 KRJ97257.1 KRT03723.1 KMY88042.1 KRT03725.1 AGB92555.1 DS235331 EEB14757.1
RSAL01000116 RVE46867.1 PCG66305.1 KQ459592 KPI96763.1 KQ460845 KPJ12031.1 ODYU01001360 SOQ37426.1 PCG66303.1 AAAB01008964 EAA12846.4 ATLV01014971 ATLV01014972 KE524999 KFB39724.1 CH477394 EAT41900.1 APCN01002500 GEZM01024234 JAV87842.1 GEZM01024223 JAV87859.1 CVRI01000044 CRK96838.1 GEZM01024232 JAV87845.1 GEZM01024228 JAV87850.1 KQ435102 KZC14664.1 GEZM01024229 JAV87849.1 GEZM01024230 JAV87848.1 AXCM01008531 UFQT01000144 SSX20849.1 GAMC01018609 JAB87946.1 GEBQ01010558 JAT29419.1 CH940661 KRF85516.1 KRF85517.1 JXUM01008362 JXUM01008363 JXUM01008364 KQ560259 KXJ83503.1 SSX20848.1 EDW71191.2 GAKP01019355 JAC39597.1 GAMC01018611 JAB87944.1 JR035185 AEY56260.1 CH963920 EDW77649.2 CM000157 KRJ97262.1 CM002910 KMY88043.1 CH379059 KRT03730.1 AJ312132 AJ312133 AJ312134 AJ312135 AE014134 BT010247 GAMC01018610 JAB87945.1 KQ971338 KYB28097.1 CP012523 ALC38083.1 KRJ97260.1 CH480820 EDW54219.1 ACZ94160.1 KMY88039.1 KRT03731.1 KRT03722.1 KYB28096.1 KRJ97258.1 OUUW01000004 SPP79099.1 ACZ94159.1 AGB92556.1 GEDC01019570 JAS17728.1 GBXI01004372 JAD09920.1 CH902620 KPU73026.1 CH916368 EDW04280.1 GEDC01010089 JAS27209.1 GBXI01008828 JAD05464.1 KRJ97264.1 CH954177 KQS70076.1 ADMH02001548 ETN62103.1 KMY88040.1 KRT03728.1 ACPB03018591 KQS70081.1 KPU73025.1 GEDC01002875 JAS34423.1 GEDC01030374 GEDC01023640 GEDC01014706 JAS06924.1 JAS13658.1 JAS22592.1 GBXI01008427 JAD05865.1 KRJ97256.1 KQS70082.1 KQS70078.1 GBYB01003570 JAG73337.1 GBYB01014906 JAG84673.1 EDW87620.1 KRJ97254.1 KRJ97255.1 KRJ97259.1 KRJ97257.1 KRT03723.1 KMY88042.1 KRT03725.1 AGB92555.1 DS235331 EEB14757.1
Proteomes
UP000218220
UP000007151
UP000283053
UP000053268
UP000053240
UP000007062
+ More
UP000030765 UP000008820 UP000075885 UP000075880 UP000069272 UP000075840 UP000075884 UP000183832 UP000075903 UP000075900 UP000076407 UP000076408 UP000076502 UP000075883 UP000008792 UP000069940 UP000249989 UP000007798 UP000002282 UP000001819 UP000000803 UP000007266 UP000092553 UP000001292 UP000268350 UP000192221 UP000007801 UP000001070 UP000008711 UP000000673 UP000015103 UP000009046
UP000030765 UP000008820 UP000075885 UP000075880 UP000069272 UP000075840 UP000075884 UP000183832 UP000075903 UP000075900 UP000076407 UP000076408 UP000076502 UP000075883 UP000008792 UP000069940 UP000249989 UP000007798 UP000002282 UP000001819 UP000000803 UP000007266 UP000092553 UP000001292 UP000268350 UP000192221 UP000007801 UP000001070 UP000008711 UP000000673 UP000015103 UP000009046
Interpro
IPR013106
Ig_V-set
+ More
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR013783 Ig-like_fold
IPR003961 FN3_dom
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR036116 FN3_sf
IPR013098 Ig_I-set
IPR013818 Lipase/vitellogenin
IPR029058 AB_hydrolase
IPR033906 Lipase_N
IPR000734 TAG_lipase
IPR024160 BIN3_SAM-bd_dom
IPR029063 SAM-dependent_MTases
IPR010675 Bin3_C
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR013783 Ig-like_fold
IPR003961 FN3_dom
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR036116 FN3_sf
IPR013098 Ig_I-set
IPR013818 Lipase/vitellogenin
IPR029058 AB_hydrolase
IPR033906 Lipase_N
IPR000734 TAG_lipase
IPR024160 BIN3_SAM-bd_dom
IPR029063 SAM-dependent_MTases
IPR010675 Bin3_C
Gene 3D
ProteinModelPortal
A0A1U9XRB3
A0A2A4J3J1
A0A212F3G4
A0A3S2NX96
A0A2A4J2M0
A0A194PUV0
+ More
A0A194R837 A0A2H1V9G2 A0A2A4J4G3 Q7Q3K8 A0A1S4J7Q9 A0A1S4GWQ1 A0A1S4FDQ5 A0A084VP30 Q0IF39 A0A182PJ73 A0A182JK20 A0A182F4L4 A0A182HV77 A0A1Y1MQA9 A0A182NE36 A0A1Y1MQ97 A0A1J1ID06 A0A182UV49 A0A182RBL8 A0A1Y1MTP8 A0A1Y1MTN2 A0A182WT84 A0A182YGU3 A0A154PTC6 A0A1Y1MQ85 A0A1Y1MRZ4 A0A182M3Y9 A0A336LVW2 W8B3X5 A0A1B6M0G0 A0A0Q9WKY0 A0A0Q9WL38 A0A182G7G0 A0A336M4C3 B4MDD0 A0A034VBV9 W8BDV1 V9I653 B4N0N6 A0A0R1DK60 A0A0J9QVH7 A0A0R3NRK4 Q967D7-5 W8AHL1 A0A139WJV7 A0A0M3QT07 A0A0R1DJ34 B4I374 Q967D7-2 E1JHS9 Q967D7-4 A0A0J9QWH3 A0A0R3NY41 A0A0R3NRT6 A0A139WJK5 A0A0R1DJ77 A0A3B0JZJ0 Q967D7 E1JHT0 A0A1B6CWC2 A0A1W4UQ05 A0A1W4V3Y0 A0A0A1XG79 A0A0P9A3B3 B4JD36 A0A1B6DNG5 A0A1W4UQJ1 A0A0A1X297 A0A0R1DQD2 A0A0Q5W9B6 A0A1W4UQI6 A0A1W4V2S8 W5JFI5 A0A1W4V2G1 A0A0J9TF92 A0A0R3NXQ3 T1HFR6 A0A0Q5W961 A0A0P8XE88 A0A1B6E919 A0A1B6C0M6 A0A0A1X554 A0A0R1DJD6 A0A0Q5WKV0 A0A0Q5WID7 A0A0C9R6F7 A0A0C9S214 B4NYI7 A0A0R1DQN2 A0A0R3NRN3 A0A0J9QVG8 A0A0R3NWW3 M9PCI8 E0VN01
A0A194R837 A0A2H1V9G2 A0A2A4J4G3 Q7Q3K8 A0A1S4J7Q9 A0A1S4GWQ1 A0A1S4FDQ5 A0A084VP30 Q0IF39 A0A182PJ73 A0A182JK20 A0A182F4L4 A0A182HV77 A0A1Y1MQA9 A0A182NE36 A0A1Y1MQ97 A0A1J1ID06 A0A182UV49 A0A182RBL8 A0A1Y1MTP8 A0A1Y1MTN2 A0A182WT84 A0A182YGU3 A0A154PTC6 A0A1Y1MQ85 A0A1Y1MRZ4 A0A182M3Y9 A0A336LVW2 W8B3X5 A0A1B6M0G0 A0A0Q9WKY0 A0A0Q9WL38 A0A182G7G0 A0A336M4C3 B4MDD0 A0A034VBV9 W8BDV1 V9I653 B4N0N6 A0A0R1DK60 A0A0J9QVH7 A0A0R3NRK4 Q967D7-5 W8AHL1 A0A139WJV7 A0A0M3QT07 A0A0R1DJ34 B4I374 Q967D7-2 E1JHS9 Q967D7-4 A0A0J9QWH3 A0A0R3NY41 A0A0R3NRT6 A0A139WJK5 A0A0R1DJ77 A0A3B0JZJ0 Q967D7 E1JHT0 A0A1B6CWC2 A0A1W4UQ05 A0A1W4V3Y0 A0A0A1XG79 A0A0P9A3B3 B4JD36 A0A1B6DNG5 A0A1W4UQJ1 A0A0A1X297 A0A0R1DQD2 A0A0Q5W9B6 A0A1W4UQI6 A0A1W4V2S8 W5JFI5 A0A1W4V2G1 A0A0J9TF92 A0A0R3NXQ3 T1HFR6 A0A0Q5W961 A0A0P8XE88 A0A1B6E919 A0A1B6C0M6 A0A0A1X554 A0A0R1DJD6 A0A0Q5WKV0 A0A0Q5WID7 A0A0C9R6F7 A0A0C9S214 B4NYI7 A0A0R1DQN2 A0A0R3NRN3 A0A0J9QVG8 A0A0R3NWW3 M9PCI8 E0VN01
PDB
3B43
E-value=9.01792e-29,
Score=319
Ontologies
GO
GO:0016021
GO:0006629
GO:0005576
GO:0052689
GO:0008168
GO:0048814
GO:0070593
GO:0007629
GO:0007411
GO:0016199
GO:0007414
GO:0008039
GO:0030537
GO:0098632
GO:0045467
GO:0005887
GO:0007638
GO:0008344
GO:0005515
GO:0046856
GO:0000166
GO:0006412
GO:0005852
GO:0006446
GO:0043022
GO:0005488
GO:0003707
GO:0051537
Topology
Subcellular location
Secreted
Membrane
Membrane
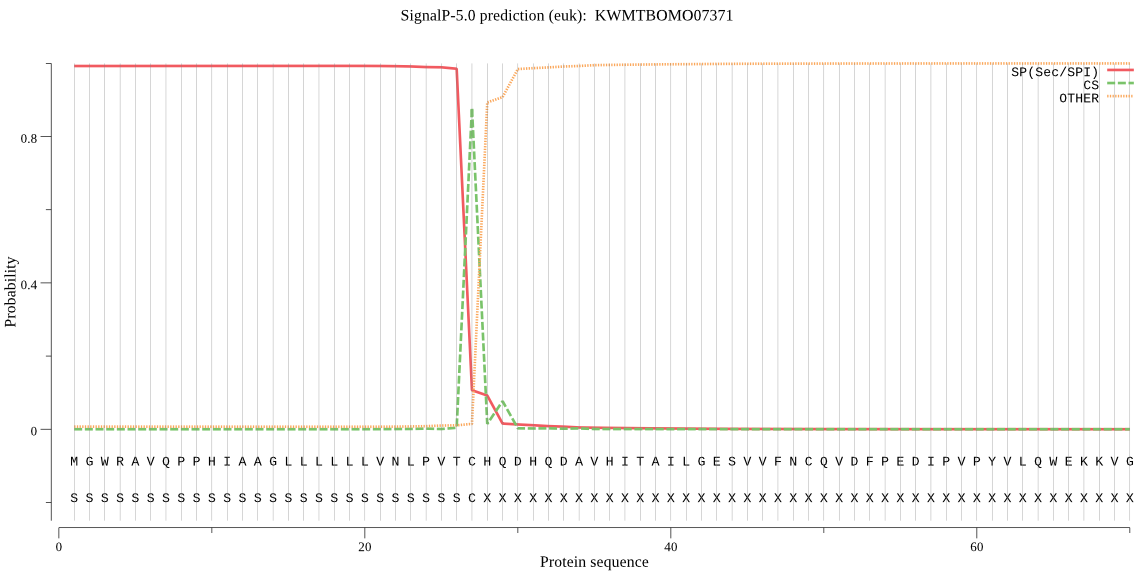
SignalP
Position: 1 - 27,
Likelihood: 0.992985
Length:
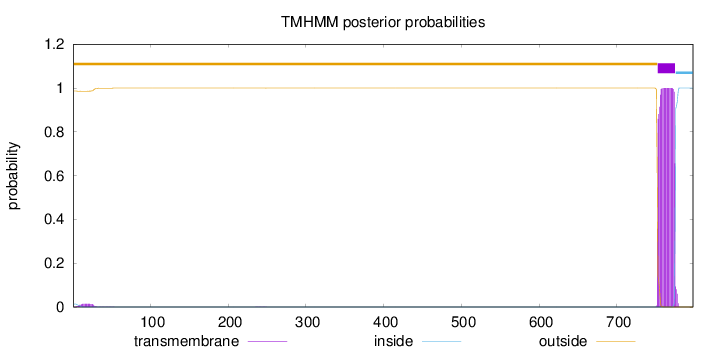
798
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.15249
Exp number, first 60 AAs:
0.28583
Total prob of N-in:
0.01389
outside
1 - 752
TMhelix
753 - 775
inside
776 - 798
Population Genetic Test Statistics
Pi
22.555375
Theta
18.845785
Tajima's D
0.02776
CLR
19.270444
CSRT
0.381030948452577
Interpretation
Uncertain
