Gene
KWMTBOMO07370
Pre Gene Modal
BGIBMGA010602
Annotation
PREDICTED:_polyphosphoinositide_phosphatase_isoform_X1_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 1.975
Sequence
CDS
ATGTCGAAAGAAAGTTTAATGTTTCATCCTATAATTAGCTCAATACAACGAATAGCGCTTTATGAAACAAAAGCTAGATTCTATTTGATTGGATCAAACAACACTGAAACGAGGTTTAGAGTTTTAAAAATTGATAGGACAGATCCAAGAGATTTGACATTAGTAGATGACAAAGTAGAGTATAACAAACAAGAAATTCATCAACTTTTGAACATGATTGGTTTTGGAAACAGAACTGGAAGATCAACAGGACTCAATAAACTAGTGTCAGCATTTGGGATTGTAGGTTTCATAAGATTCTTAGAGGGCTATTATATTATACTAGTCACAAAACGTCGGAAGATCGCAGCTATTGGTTCACATTCCATTTACAAAATAGAAGATACAGCAACGATATACATTCCGAATGAAAACAATAAGCCTCCTCATCCTGATGAACATCGCTACCTCAAAATGTTCTTAGCTATTGACTTGTCCACCAACTTTTACTACAGCTATAGTTATGATATCACTCACACATTGCAAATGAACATGGCACCTCCAAGACAACTAGCTCCGGCGTTATTCCCTAAACCCATCACAGCAGCAGTATATCAATCAAACTTGAACACTGATAAAGCAGGTGACATGGGATGTGAATGTAGAAAAAAAGAAGACGATGAAGACATTTTTGAAACTTGGAAGAAACAAATTCGACAAAGACAAGAAAAGACAGGAGGAAAAAGTCCAAAGCTAGAATTCGGAGTGCGCAGTGTACCGGAGTGGAGGTTTGTTTGGAACAGTCACCTGCTATCTCCAGTCCACTCACATCTGCATCCAGATTGGATATTGTACATTGTTCATGGGTTCATTGAACAAAGTAATCTAAATATATTCGGAAGAGCTGTATATCTCACTCTTATAGCCAGAAGGAGTAACAGATACGCTGGTACTAGATTCCTTAAGAGAGGTGCTAATATGCATGGAGATGTCGCAAATGAAGTGGAGACTGAGCAAATTGTACATGATTCAATGGTATCATCTTTTGCAGCTGGAAGATTTAGCTCATTTGTACAAATGAGAGGTTCAATACCAAGTTACTGGTCACAAGACATCTCGAAGATGGTACCCAAGCCAGCAATATCGATTGACCTCGGAGATCCGTTTGCAGAAATACCAGGAAAACATTTGAACAATTTAATGCGAAGATATGGGTCTCCAATAATGTTCCTAAATTTAGTTAAGAAAAGGGAAAAGAAAAAACACGAATCTTTATTGACCGATGTGATTAGTAACGGCATCAAATATTTGAATCAGTTCTTACCCCCTGAGCATGCAATACAATATTATCATTTAGACATGGCTAGAGTCAACAAGGGCTCCGAGGCAAAGGTTTTGGACAAATTATCGGCTATAGCCCGTACAGTAGTGCGTAAAACTGGTATATTTTTGAGTTTAAATGGCTACGGAGAAGAGATCGGTGTTGAGGGGCATCAGAGCGGTCGCTTACAAACCGGTCTTGTTCGAGTCAACTGCGTGGATTGCCTCGACAGAACGAACACGGCTCAGTTTGCAATCGGAAAATGCGTACTAGCTTATCAGCTATATGCTTTAGGTTTGATCGGTACACCGGTTATAGAATTCGACTCGGACTGTGCGAGGCTACTTGAAGCTTTATATGAAGACCACGGAGACACACTAGCTCTGCAGTATGGCGGCTCCCAACTTGTACATAGAATAAAAACGTACCGCAAGACAGCTCCGTGGTCCTCGCACGGCAACGACATAATGCAGACCCTTAGCCGGTATTACTCGAACACGTTCTCGGATGCCGAGAAACAGAACACCATGAATTTGTTCCTGGGCCTATTCATACCGGACCCGACGAAGCATCCCATCTGGGAGTTCCCGACCGATCACTACCTGCATCATCCTGAAGCATCGGTGTACATACCGAGGACTAGGTGCGTAACATATTCCTAA
Protein
MSKESLMFHPIISSIQRIALYETKARFYLIGSNNTETRFRVLKIDRTDPRDLTLVDDKVEYNKQEIHQLLNMIGFGNRTGRSTGLNKLVSAFGIVGFIRFLEGYYIILVTKRRKIAAIGSHSIYKIEDTATIYIPNENNKPPHPDEHRYLKMFLAIDLSTNFYYSYSYDITHTLQMNMAPPRQLAPALFPKPITAAVYQSNLNTDKAGDMGCECRKKEDDEDIFETWKKQIRQRQEKTGGKSPKLEFGVRSVPEWRFVWNSHLLSPVHSHLHPDWILYIVHGFIEQSNLNIFGRAVYLTLIARRSNRYAGTRFLKRGANMHGDVANEVETEQIVHDSMVSSFAAGRFSSFVQMRGSIPSYWSQDISKMVPKPAISIDLGDPFAEIPGKHLNNLMRRYGSPIMFLNLVKKREKKKHESLLTDVISNGIKYLNQFLPPEHAIQYYHLDMARVNKGSEAKVLDKLSAIARTVVRKTGIFLSLNGYGEEIGVEGHQSGRLQTGLVRVNCVDCLDRTNTAQFAIGKCVLAYQLYALGLIGTPVIEFDSDCARLLEALYEDHGDTLALQYGGSQLVHRIKTYRKTAPWSSHGNDIMQTLSRYYSNTFSDAEKQNTMNLFLGLFIPDPTKHPIWEFPTDHYLHHPEASVYIPRTRCVTYS
Summary
Uniprot
H9JM50
A0A194PHA8
A0A2H1VAR9
A0A2A4JE40
A0A194R2L5
A0A1E1WI33
+ More
A0A3S2TIF6 A0A212F3G2 A0A067QVE8 A0A2J7PQ51 A0A2J7PQ53 A0A1B6EWM8 A0A0C9QXJ0 A0A2J7PQ55 A0A1B6JCG7 A0A2J7PQ52 A0A2J7PQ95 D2A1F9 A0A0T6AZD2 A0A195BVF8 F4W5F5 A0A0M8ZXJ3 A0A195D0U4 A0A154PTJ0 A0A0L7RDT7 A0A1Y1MFK9 A0A026X451 A0A1B6C3M9 A0A195FSR7 A0A151WQH3 A0A1B6BYX7 E9GGD2 A0A0N7ZKX1 A0A1W4XVC6 A0A224X7B4 A0A1E1XCT7 A0A1L8G9D7 C3YI49 A0A2J8U1I0 A0A099ZPA1 G3RI08 A0A2K5D7Q8 A0A2K5QFW2 A0A2K6TA35 K7DGN1 A0A2K6EBU4 A0A2K5XGF9 A0A0D9RSD2 A0A096MVE6 A0A2K5L7N2 G7P3N7 G7MQX2 V9KFY4 A0A2K6PS44 F7D8B2 U3FF16 A0A2R8ZFV1 A0A2K6PS42 A0A2K5L7P9 U3CMT1 A0A250Y5V9 A0A3Q0EI22 F6UNT5 A0A131Z056 A0A224Z526 L7MLL8 A0A2Y9QJ17 A0A093QJP0 V8P7F6 A0A2Y9DDS3 A0A3Q3AL80 A0A091GAV4 A0A1V4JN98 G3VVI9 G1KFD9 F7E3B1 E0VXU4 A0A091LQQ8 H0ZPC1 H2PK13 A0A218UYV7 A0A384DBP3 V5HZE8 A0A2U9CV69 A0A3P8Q5C5 A0A3B4FVK9 A0A3Q2VBT9 A0A3P8Q6F7 A0A3P8Q4T7 A0A3P9CLM0 A0A3S2TUV1 G1NL57 A0A2S2PUL1 A0A3P9M5T0 A0A3P8S685 A0A2S2QTC3 A0A087X413 A0A3P8S7P9 A0A3B3UJB5 A0A3P8WZV2
A0A3S2TIF6 A0A212F3G2 A0A067QVE8 A0A2J7PQ51 A0A2J7PQ53 A0A1B6EWM8 A0A0C9QXJ0 A0A2J7PQ55 A0A1B6JCG7 A0A2J7PQ52 A0A2J7PQ95 D2A1F9 A0A0T6AZD2 A0A195BVF8 F4W5F5 A0A0M8ZXJ3 A0A195D0U4 A0A154PTJ0 A0A0L7RDT7 A0A1Y1MFK9 A0A026X451 A0A1B6C3M9 A0A195FSR7 A0A151WQH3 A0A1B6BYX7 E9GGD2 A0A0N7ZKX1 A0A1W4XVC6 A0A224X7B4 A0A1E1XCT7 A0A1L8G9D7 C3YI49 A0A2J8U1I0 A0A099ZPA1 G3RI08 A0A2K5D7Q8 A0A2K5QFW2 A0A2K6TA35 K7DGN1 A0A2K6EBU4 A0A2K5XGF9 A0A0D9RSD2 A0A096MVE6 A0A2K5L7N2 G7P3N7 G7MQX2 V9KFY4 A0A2K6PS44 F7D8B2 U3FF16 A0A2R8ZFV1 A0A2K6PS42 A0A2K5L7P9 U3CMT1 A0A250Y5V9 A0A3Q0EI22 F6UNT5 A0A131Z056 A0A224Z526 L7MLL8 A0A2Y9QJ17 A0A093QJP0 V8P7F6 A0A2Y9DDS3 A0A3Q3AL80 A0A091GAV4 A0A1V4JN98 G3VVI9 G1KFD9 F7E3B1 E0VXU4 A0A091LQQ8 H0ZPC1 H2PK13 A0A218UYV7 A0A384DBP3 V5HZE8 A0A2U9CV69 A0A3P8Q5C5 A0A3B4FVK9 A0A3Q2VBT9 A0A3P8Q6F7 A0A3P8Q4T7 A0A3P9CLM0 A0A3S2TUV1 G1NL57 A0A2S2PUL1 A0A3P9M5T0 A0A3P8S685 A0A2S2QTC3 A0A087X413 A0A3P8S7P9 A0A3B3UJB5 A0A3P8WZV2
Pubmed
19121390
26354079
22118469
24845553
18362917
19820115
+ More
21719571 28004739 24508170 21292972 28503490 27762356 18563158 22398555 16136131 22002653 25319552 24402279 25362486 17431167 25243066 22722832 28087693 26830274 28797301 25576852 24297900 15592404 21709235 17495919 20566863 20360741 25765539 25186727 20838655 17554307 24487278
21719571 28004739 24508170 21292972 28503490 27762356 18563158 22398555 16136131 22002653 25319552 24402279 25362486 17431167 25243066 22722832 28087693 26830274 28797301 25576852 24297900 15592404 21709235 17495919 20566863 20360741 25765539 25186727 20838655 17554307 24487278
EMBL
BABH01033517
BABH01033518
KQ459603
KPI92692.1
ODYU01001559
SOQ37943.1
+ More
NWSH01001812 PCG70046.1 KQ460845 KPJ12033.1 GDQN01004400 JAT86654.1 RSAL01000116 RVE46868.1 AGBW02010564 OWR48279.1 KK852904 KDR14022.1 NEVH01022640 PNF18467.1 PNF18469.1 GECZ01027427 JAS42342.1 GBYB01000395 JAG70162.1 PNF18468.1 GECU01010812 JAS96894.1 PNF18463.1 PNF18466.1 KQ971338 EFA02109.1 LJIG01022504 KRT80221.1 KQ976407 KYM91465.1 GL887645 EGI70605.1 KQ435824 KOX72106.1 KQ977004 KYN06533.1 KQ435102 KZC14668.1 KQ414613 KOC69018.1 GEZM01033041 JAV84453.1 KK107013 EZA62883.1 GEDC01029187 JAS08111.1 KQ981276 KYN43635.1 KQ982821 KYQ50149.1 GEDC01030827 JAS06471.1 GL732543 EFX81517.1 GDIP01235369 JAI88032.1 GFTR01008130 JAW08296.1 GFAC01002121 JAT97067.1 CM004474 OCT80385.1 GG666514 EEN60186.1 NDHI03003476 PNJ39129.1 KL897257 KGL84239.1 CABD030047212 CABD030047213 CABD030047214 CABD030047215 CABD030047216 CABD030047217 CABD030047218 CABD030047219 CABD030047220 AACZ04054319 AACZ04054320 AACZ04054321 AACZ04054322 GABC01006721 GABF01007132 GABD01008456 GABE01011507 NBAG03000210 JAA04617.1 JAA15013.1 JAA24644.1 JAA33232.1 PNI87644.1 AQIB01069387 AQIB01069388 AQIB01069389 AQIB01069390 AQIB01069391 AQIB01069392 AQIB01069393 AQIB01069394 AQIB01069395 AQIB01069396 AHZZ02025978 AHZZ02025979 AQIA01049399 AQIA01049400 CM001279 EHH53463.1 JU319900 CM001256 AFE63656.1 EHH18783.1 JW864190 AFO96707.1 JSUE03032505 JSUE03032506 GAMT01008919 GAMQ01006236 JAB02942.1 JAB35615.1 AJFE02014513 AJFE02014514 AJFE02014515 AJFE02014516 GAMS01009393 GAMR01006904 GAMP01008111 JAB13743.1 JAB27028.1 JAB44644.1 GFFV01000960 JAV38985.1 GEDV01004701 JAP83856.1 GFPF01010587 MAA21733.1 GACK01000975 JAA64059.1 KL672036 KFW84242.1 AZIM01000655 ETE69943.1 AADN05000003 AC145954 KL447977 KFO79098.1 LSYS01006902 OPJ73197.1 AEFK01170552 AEFK01170553 AEFK01170554 AEFK01170555 AEFK01170556 AEFK01170557 AEFK01170558 AEFK01170559 AEFK01170560 DS235840 EEB18200.1 KK504099 KFP60719.1 ABQF01000415 ABGA01136125 ABGA01136126 ABGA01136127 ABGA01162126 ABGA01162127 ABGA01162128 ABGA01162129 ABGA01273179 ABGA01273180 ABGA01273181 MUZQ01000087 OWK59005.1 GANP01002526 JAB81942.1 CP026262 AWP20053.1 CM012460 RVE55803.1 GGMR01020395 MBY33014.1 GGMS01011816 MBY81019.1 AYCK01005070 AYCK01005071
NWSH01001812 PCG70046.1 KQ460845 KPJ12033.1 GDQN01004400 JAT86654.1 RSAL01000116 RVE46868.1 AGBW02010564 OWR48279.1 KK852904 KDR14022.1 NEVH01022640 PNF18467.1 PNF18469.1 GECZ01027427 JAS42342.1 GBYB01000395 JAG70162.1 PNF18468.1 GECU01010812 JAS96894.1 PNF18463.1 PNF18466.1 KQ971338 EFA02109.1 LJIG01022504 KRT80221.1 KQ976407 KYM91465.1 GL887645 EGI70605.1 KQ435824 KOX72106.1 KQ977004 KYN06533.1 KQ435102 KZC14668.1 KQ414613 KOC69018.1 GEZM01033041 JAV84453.1 KK107013 EZA62883.1 GEDC01029187 JAS08111.1 KQ981276 KYN43635.1 KQ982821 KYQ50149.1 GEDC01030827 JAS06471.1 GL732543 EFX81517.1 GDIP01235369 JAI88032.1 GFTR01008130 JAW08296.1 GFAC01002121 JAT97067.1 CM004474 OCT80385.1 GG666514 EEN60186.1 NDHI03003476 PNJ39129.1 KL897257 KGL84239.1 CABD030047212 CABD030047213 CABD030047214 CABD030047215 CABD030047216 CABD030047217 CABD030047218 CABD030047219 CABD030047220 AACZ04054319 AACZ04054320 AACZ04054321 AACZ04054322 GABC01006721 GABF01007132 GABD01008456 GABE01011507 NBAG03000210 JAA04617.1 JAA15013.1 JAA24644.1 JAA33232.1 PNI87644.1 AQIB01069387 AQIB01069388 AQIB01069389 AQIB01069390 AQIB01069391 AQIB01069392 AQIB01069393 AQIB01069394 AQIB01069395 AQIB01069396 AHZZ02025978 AHZZ02025979 AQIA01049399 AQIA01049400 CM001279 EHH53463.1 JU319900 CM001256 AFE63656.1 EHH18783.1 JW864190 AFO96707.1 JSUE03032505 JSUE03032506 GAMT01008919 GAMQ01006236 JAB02942.1 JAB35615.1 AJFE02014513 AJFE02014514 AJFE02014515 AJFE02014516 GAMS01009393 GAMR01006904 GAMP01008111 JAB13743.1 JAB27028.1 JAB44644.1 GFFV01000960 JAV38985.1 GEDV01004701 JAP83856.1 GFPF01010587 MAA21733.1 GACK01000975 JAA64059.1 KL672036 KFW84242.1 AZIM01000655 ETE69943.1 AADN05000003 AC145954 KL447977 KFO79098.1 LSYS01006902 OPJ73197.1 AEFK01170552 AEFK01170553 AEFK01170554 AEFK01170555 AEFK01170556 AEFK01170557 AEFK01170558 AEFK01170559 AEFK01170560 DS235840 EEB18200.1 KK504099 KFP60719.1 ABQF01000415 ABGA01136125 ABGA01136126 ABGA01136127 ABGA01162126 ABGA01162127 ABGA01162128 ABGA01162129 ABGA01273179 ABGA01273180 ABGA01273181 MUZQ01000087 OWK59005.1 GANP01002526 JAB81942.1 CP026262 AWP20053.1 CM012460 RVE55803.1 GGMR01020395 MBY33014.1 GGMS01011816 MBY81019.1 AYCK01005070 AYCK01005071
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000283053
UP000007151
+ More
UP000027135 UP000235965 UP000007266 UP000078540 UP000007755 UP000053105 UP000078542 UP000076502 UP000053825 UP000053097 UP000078541 UP000075809 UP000000305 UP000192223 UP000186698 UP000001554 UP000053641 UP000001519 UP000233020 UP000233040 UP000233220 UP000002277 UP000233120 UP000233140 UP000029965 UP000028761 UP000233060 UP000009130 UP000233100 UP000233200 UP000006718 UP000240080 UP000008225 UP000189704 UP000248480 UP000053258 UP000000539 UP000053760 UP000190648 UP000007648 UP000001646 UP000002280 UP000009046 UP000007754 UP000001595 UP000197619 UP000261680 UP000246464 UP000265100 UP000261460 UP000264840 UP000265160 UP000001645 UP000265180 UP000265080 UP000028760 UP000261500 UP000265120
UP000027135 UP000235965 UP000007266 UP000078540 UP000007755 UP000053105 UP000078542 UP000076502 UP000053825 UP000053097 UP000078541 UP000075809 UP000000305 UP000192223 UP000186698 UP000001554 UP000053641 UP000001519 UP000233020 UP000233040 UP000233220 UP000002277 UP000233120 UP000233140 UP000029965 UP000028761 UP000233060 UP000009130 UP000233100 UP000233200 UP000006718 UP000240080 UP000008225 UP000189704 UP000248480 UP000053258 UP000000539 UP000053760 UP000190648 UP000007648 UP000001646 UP000002280 UP000009046 UP000007754 UP000001595 UP000197619 UP000261680 UP000246464 UP000265100 UP000261460 UP000264840 UP000265160 UP000001645 UP000265180 UP000265080 UP000028760 UP000261500 UP000265120
PRIDE
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
H9JM50
A0A194PHA8
A0A2H1VAR9
A0A2A4JE40
A0A194R2L5
A0A1E1WI33
+ More
A0A3S2TIF6 A0A212F3G2 A0A067QVE8 A0A2J7PQ51 A0A2J7PQ53 A0A1B6EWM8 A0A0C9QXJ0 A0A2J7PQ55 A0A1B6JCG7 A0A2J7PQ52 A0A2J7PQ95 D2A1F9 A0A0T6AZD2 A0A195BVF8 F4W5F5 A0A0M8ZXJ3 A0A195D0U4 A0A154PTJ0 A0A0L7RDT7 A0A1Y1MFK9 A0A026X451 A0A1B6C3M9 A0A195FSR7 A0A151WQH3 A0A1B6BYX7 E9GGD2 A0A0N7ZKX1 A0A1W4XVC6 A0A224X7B4 A0A1E1XCT7 A0A1L8G9D7 C3YI49 A0A2J8U1I0 A0A099ZPA1 G3RI08 A0A2K5D7Q8 A0A2K5QFW2 A0A2K6TA35 K7DGN1 A0A2K6EBU4 A0A2K5XGF9 A0A0D9RSD2 A0A096MVE6 A0A2K5L7N2 G7P3N7 G7MQX2 V9KFY4 A0A2K6PS44 F7D8B2 U3FF16 A0A2R8ZFV1 A0A2K6PS42 A0A2K5L7P9 U3CMT1 A0A250Y5V9 A0A3Q0EI22 F6UNT5 A0A131Z056 A0A224Z526 L7MLL8 A0A2Y9QJ17 A0A093QJP0 V8P7F6 A0A2Y9DDS3 A0A3Q3AL80 A0A091GAV4 A0A1V4JN98 G3VVI9 G1KFD9 F7E3B1 E0VXU4 A0A091LQQ8 H0ZPC1 H2PK13 A0A218UYV7 A0A384DBP3 V5HZE8 A0A2U9CV69 A0A3P8Q5C5 A0A3B4FVK9 A0A3Q2VBT9 A0A3P8Q6F7 A0A3P8Q4T7 A0A3P9CLM0 A0A3S2TUV1 G1NL57 A0A2S2PUL1 A0A3P9M5T0 A0A3P8S685 A0A2S2QTC3 A0A087X413 A0A3P8S7P9 A0A3B3UJB5 A0A3P8WZV2
A0A3S2TIF6 A0A212F3G2 A0A067QVE8 A0A2J7PQ51 A0A2J7PQ53 A0A1B6EWM8 A0A0C9QXJ0 A0A2J7PQ55 A0A1B6JCG7 A0A2J7PQ52 A0A2J7PQ95 D2A1F9 A0A0T6AZD2 A0A195BVF8 F4W5F5 A0A0M8ZXJ3 A0A195D0U4 A0A154PTJ0 A0A0L7RDT7 A0A1Y1MFK9 A0A026X451 A0A1B6C3M9 A0A195FSR7 A0A151WQH3 A0A1B6BYX7 E9GGD2 A0A0N7ZKX1 A0A1W4XVC6 A0A224X7B4 A0A1E1XCT7 A0A1L8G9D7 C3YI49 A0A2J8U1I0 A0A099ZPA1 G3RI08 A0A2K5D7Q8 A0A2K5QFW2 A0A2K6TA35 K7DGN1 A0A2K6EBU4 A0A2K5XGF9 A0A0D9RSD2 A0A096MVE6 A0A2K5L7N2 G7P3N7 G7MQX2 V9KFY4 A0A2K6PS44 F7D8B2 U3FF16 A0A2R8ZFV1 A0A2K6PS42 A0A2K5L7P9 U3CMT1 A0A250Y5V9 A0A3Q0EI22 F6UNT5 A0A131Z056 A0A224Z526 L7MLL8 A0A2Y9QJ17 A0A093QJP0 V8P7F6 A0A2Y9DDS3 A0A3Q3AL80 A0A091GAV4 A0A1V4JN98 G3VVI9 G1KFD9 F7E3B1 E0VXU4 A0A091LQQ8 H0ZPC1 H2PK13 A0A218UYV7 A0A384DBP3 V5HZE8 A0A2U9CV69 A0A3P8Q5C5 A0A3B4FVK9 A0A3Q2VBT9 A0A3P8Q6F7 A0A3P8Q4T7 A0A3P9CLM0 A0A3S2TUV1 G1NL57 A0A2S2PUL1 A0A3P9M5T0 A0A3P8S685 A0A2S2QTC3 A0A087X413 A0A3P8S7P9 A0A3B3UJB5 A0A3P8WZV2
PDB
4TU3
E-value=4.79864e-31,
Score=338
Ontologies
PATHWAY
GO
PANTHER
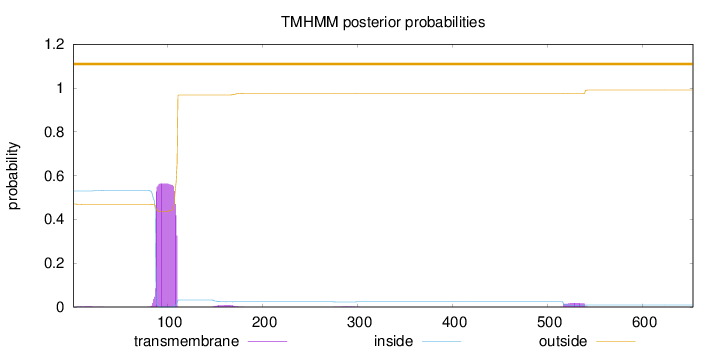
Topology
Length:
653
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
13.18887
Exp number, first 60 AAs:
0.04668
Total prob of N-in:
0.52955
outside
1 - 653
Population Genetic Test Statistics
Pi
1.378096
Theta
5.506687
Tajima's D
-2.34359
CLR
637.706737
CSRT
0.00149992500374981
Interpretation
Possibly Positive selection
