Gene
KWMTBOMO07361
Annotation
hypothetical_protein_[Piscirickettsia_salmonis]
Location in the cell
Mitochondrial Reliability : 2.185
Sequence
CDS
ATGGTCCCCCCCCCGGCGATGGTGACCAGGGGCCCCGGGATCGTCGCGGTACAATTAGAGAATATCGTCGTGATCGGGGTGTACTTTTCTCCGAATCGGCCTACCGTCGAGTTCCAGCGGTTCCTGGACGGGCTAGAGGTGATCGCTCGTCAACTCGCTCCCCGTTCAGTGATACTGGCGGGGGACTTCAATGCGAAGTCTGTCGCTTGGGGTTCCCCATTCACGGACGCTCGTGGTAGGCTGCTGGAGGAGTGGGCGGTCGCGGTCGGTCTCTGTATCGTTAATAGGGGCTCGATCGCGACTTGCGTGCGGTGGACGGGCGAGTCTATCGTGGACTTGACGTTCGCGAGCTCGCCCGTCGCACGGCGCATCCTCGGCTGGAGAGTGGTGGTGGGGGCGGAATCGTTGTCCGATCACCGGTATATCCGGTACGATCTTTCTGCCCCCGCGCCGACGCCGGCTGCAGGTGCCCGCGGGGTTGACCCCCTGCAGAGTGCATCCCGGTCATTCCCTAGGTGGGCACTGAAGCTCCTGAACAGGGAGCTCCTGGTGGAGGCCTCTATGGTGGCGGCGTGGGCGCCCAAGCGCCCACAACCTGTCGATGTGGAAACCGAGGCCGAATGGTTCCGGGGCGCGATGCACCGCATATGTGATGCCTCGATGCCCCGGGTAGGCCCTCGGCATTCTGGACGGCGACAGTCGCCCTGGTGGTCGCCCGAAATTGCAAGACTGCGTGCGGTCTCCATTCGGGCGAGCCGCCAGTGCACTAGACACCGCCGTCGCCGCCGCCTGCGGCGCGACGAGCCTGTCGCGTTCGCGGAGGAGGAAGCTCGGCTGCACCGCGAATATCGCGCTGCGAAGGACGCCCTGCGGCTGGCCATCAGGCGGGCCAAGGAGCAGAACATGGAGAGACTCTTGGAGGCGCTCGAAGCGGATCCGTGGGGGAGCCCATACAAATTGGTACGCGGCAAGTTACGCCCGTGGGCCCCCTCTATGACTGAGCGTCTCCAGCCTCAGCAGCTGCGGGACATCGTTTCGGCGTTGTTCCCGCAGGAGCGGGAGGGTTTTGTCCCTCCCGCTATGGGCGGGCCTTCTGACACCGTCGACGGCCAAGCTCCTGCTGAGGTGCCCCGTATTACCGAGGCAGAGCTCCGGGTGGCCGTTGTCAAGATGACTGCGAAAGACACGGCCCCCGGCCCAGACGGAGTCCACGGCCGGGTATTGGCCTTGGCCCTCGGTGCCCTGGGGGACCGGCTCCTTGAGCTATATAATGGCTGCTTGGAGTCGGGACGGTTTCCGTCGCTCTGGCGGACGGGTAGACTTGTGTTGTTGAGGAAGGAGGGGCGCCCGGTGGATACAGCCGCCGGGTATCGTCCTATCGTACTGCTGGACGAGGCGGGAAAATTGCTGGAACGTATTCTGGCTGCCCGCATCGTTCAGCACCTGGTCGGGGTGGGGCCTGACCTGTCGGCGGAGCAGTACGGCTTCCGGGAGGGCCGTTCGACCGTGGATGCAATTCTTCGCGTGCGGTCCCTCTCGGACGAGGCTGTTTCTCGGGGTGGGGTGGCGCTGGCGGTGTCTCTTGACATCGCCAACGCATTTAACACTCTGTCCTGGACCGTGATAGGGGGGGCACTGGAGAGGCATGGAGTGCCCCTCTACCTCCGCCGGCTGCGGGAGTGGCCCTCGTCGTCGGAAGGATAA
Protein
MVPPPAMVTRGPGIVAVQLENIVVIGVYFSPNRPTVEFQRFLDGLEVIARQLAPRSVILAGDFNAKSVAWGSPFTDARGRLLEEWAVAVGLCIVNRGSIATCVRWTGESIVDLTFASSPVARRILGWRVVVGAESLSDHRYIRYDLSAPAPTPAAGARGVDPLQSASRSFPRWALKLLNRELLVEASMVAAWAPKRPQPVDVETEAEWFRGAMHRICDASMPRVGPRHSGRRQSPWWSPEIARLRAVSIRASRQCTRHRRRRRLRRDEPVAFAEEEARLHREYRAAKDALRLAIRRAKEQNMERLLEALEADPWGSPYKLVRGKLRPWAPSMTERLQPQQLRDIVSALFPQEREGFVPPAMGGPSDTVDGQAPAEVPRITEAELRVAVVKMTAKDTAPGPDGVHGRVLALALGALGDRLLELYNGCLESGRFPSLWRTGRLVLLRKEGRPVDTAAGYRPIVLLDEAGKLLERILAARIVQHLVGVGPDLSAEQYGFREGRSTVDAILRVRSLSDEAVSRGGVALAVSLDIANAFNTLSWTVIGGALERHGVPLYLRRLREWPSSSEG
Summary
Uniprot
Q868Q4
A0A212EYI2
Q8MY29
Q8MY37
Q8MY31
Q8MY35
+ More
Q8MY33 Q8MY25 Q8MY27 A0A0J7N1D9 O01419 A0A2W1C3J5 A0A212FI97 E2AA34 A0A0J7KJG6 A0A0J7KF37 A0A0J7K9N6 A0A0J7KQK3 A0A0J7KIY5 A0A0J7KIM6 A0A0J7N0R3 A0A0J7KIC2 A0A0J7KFH2 A0A0J7NC28 A0A0J7KQY4 A0A0J7MY17 A0A0J7NKG1 A0A0J7KCE1 A0A0J7KF91 A0A0J7NFZ0 A0A0J7MWN5 A0A0J7KRB8 A0A0J7K3F1 A0A0J7KYD2 A0A3S2LNH8 A0A0J7KIS5 A0A2S2PEZ7 A0A0J7KPR0 A0A2S2NQQ6 A0A0J7KDK4 A0A0J7K903 J9KNN8 A0A2S2PRL8 A0A0J7KF40 A0A0J7KJH4 J9LA43 J9K6F0 A0A0J7KLS2 E2BP55 J9LVU7 J9KNS1 J9L0J0 A0A0A9Z010 F7IYV5 X1X2B5 A0A0A9ZCY0 X1WJY4 A0A0J7K1J9 A0A2M4BC18 A0A2M4BC24 A0A0J7NJ13 J9LM37 J9LQQ1 J9KM84 A0A2H8TIE5 A0A0K8TR60 J9KT61 X1WZ64 J9JY94 X1WXT5 A0A0J7NAV8 J9KJG8 A0A0J7N8R4 A0A2S2PKY3 J9KUM6 A0A1S4EMG7 E0VX34 A0A2M4CL98 A0A2S2P7D1 A0A2H8TR36 A0A2M4BC28 A0A2M4BB89 A0A224XIZ2 A0A2M4BB78 A0A0J7KCD9 A0A2M4AK35 A0A2M4CS98 A0A2M3Z1V8 Q868R0 A0A142LX43 A0A2M3Z1C1 A0A0K8VPH1
Q8MY33 Q8MY25 Q8MY27 A0A0J7N1D9 O01419 A0A2W1C3J5 A0A212FI97 E2AA34 A0A0J7KJG6 A0A0J7KF37 A0A0J7K9N6 A0A0J7KQK3 A0A0J7KIY5 A0A0J7KIM6 A0A0J7N0R3 A0A0J7KIC2 A0A0J7KFH2 A0A0J7NC28 A0A0J7KQY4 A0A0J7MY17 A0A0J7NKG1 A0A0J7KCE1 A0A0J7KF91 A0A0J7NFZ0 A0A0J7MWN5 A0A0J7KRB8 A0A0J7K3F1 A0A0J7KYD2 A0A3S2LNH8 A0A0J7KIS5 A0A2S2PEZ7 A0A0J7KPR0 A0A2S2NQQ6 A0A0J7KDK4 A0A0J7K903 J9KNN8 A0A2S2PRL8 A0A0J7KF40 A0A0J7KJH4 J9LA43 J9K6F0 A0A0J7KLS2 E2BP55 J9LVU7 J9KNS1 J9L0J0 A0A0A9Z010 F7IYV5 X1X2B5 A0A0A9ZCY0 X1WJY4 A0A0J7K1J9 A0A2M4BC18 A0A2M4BC24 A0A0J7NJ13 J9LM37 J9LQQ1 J9KM84 A0A2H8TIE5 A0A0K8TR60 J9KT61 X1WZ64 J9JY94 X1WXT5 A0A0J7NAV8 J9KJG8 A0A0J7N8R4 A0A2S2PKY3 J9KUM6 A0A1S4EMG7 E0VX34 A0A2M4CL98 A0A2S2P7D1 A0A2H8TR36 A0A2M4BC28 A0A2M4BB89 A0A224XIZ2 A0A2M4BB78 A0A0J7KCD9 A0A2M4AK35 A0A2M4CS98 A0A2M3Z1V8 Q868R0 A0A142LX43 A0A2M3Z1C1 A0A0K8VPH1
EMBL
AB090825
BAC57926.1
AGBW02011529
OWR46521.1
AB078934
BAC06460.1
+ More
AB078928 BAC06450.1 AB078931 BAC06456.1 AB078929 BAC06452.1 AB078930 BAC06454.1 AB078936 BAC06464.1 AB078935 BAC06462.1 LBMM01012027 KMQ86470.1 D85594 BAA19776.1 KZ149896 PZC78733.1 AGBW02008417 OWR53447.1 GL438017 EFN69706.1 LBMM01006671 KMQ90407.1 LBMM01008448 KMQ88867.1 LBMM01011179 KMQ86989.1 LBMM01004272 KMQ92618.1 LBMM01006954 KMQ90166.1 LBMM01007036 KMQ90099.1 LBMM01012412 KMQ86235.1 LBMM01006945 KMQ90173.1 LBMM01008278 KMQ89009.1 LBMM01006996 KMQ90130.1 LBMM01004262 KMQ92649.1 LBMM01014122 KMQ85315.1 LBMM01003941 KMQ92970.1 LBMM01009535 KMQ88053.1 LBMM01008445 KMQ88871.1 LBMM01005474 KMQ91485.1 LBMM01015285 KMQ84870.1 LBMM01003988 KMQ92922.1 LBMM01015025 KMQ84953.1 LBMM01002052 KMQ95336.1 RSAL01003504 RVE40273.1 LBMM01006977 KMQ90149.1 GGMR01015159 MBY27778.1 LBMM01004634 KMQ92224.1 GGMR01006838 MBY19457.1 LBMM01008902 KMQ88543.1 LBMM01011694 KMQ86686.1 ABLF02023257 GGMR01019493 MBY32112.1 LBMM01008422 KMQ88894.1 LBMM01006657 KMQ90417.1 ABLF02018098 ABLF02012448 ABLF02012450 ABLF02012455 ABLF02012460 LBMM01005634 KMQ91348.1 GL449531 EFN82526.1 ABLF02034925 ABLF02018808 ABLF02027248 GBHO01006398 JAG37206.1 AB593325 BAK38646.1 ABLF02019402 ABLF02059310 GBHO01002406 JAG41198.1 ABLF02011238 LBMM01016720 KMQ84328.1 GGFJ01001453 MBW50594.1 GGFJ01001454 MBW50595.1 LBMM01004367 KMQ92505.1 ABLF02014650 ABLF02030663 ABLF02041312 ABLF02018942 ABLF02041885 GFXV01002090 MBW13895.1 GDAI01000741 JAI16862.1 ABLF02009073 ABLF02030702 ABLF02042963 ABLF02011183 ABLF02041884 ABLF02007989 ABLF02004854 ABLF02041317 LBMM01007425 KMQ89740.1 ABLF02041886 LBMM01008226 KMQ89040.1 GGMR01017503 MBY30122.1 ABLF02041557 DS235827 EEB17940.1 GGFL01001916 MBW66094.1 GGMR01012748 MBY25367.1 GFXV01003903 MBW15708.1 GGFJ01001439 MBW50580.1 GGFJ01001165 MBW50306.1 GFTR01008427 JAW07999.1 GGFJ01001164 MBW50305.1 LBMM01009610 KMQ87992.1 GGFK01007771 MBW41092.1 GGFL01004044 MBW68222.1 GGFM01001733 MBW22484.1 AB090822 BAC57920.1 KU543682 AMS38369.1 GGFM01001554 MBW22305.1 GDHF01029467 GDHF01011533 JAI22847.1 JAI40781.1
AB078928 BAC06450.1 AB078931 BAC06456.1 AB078929 BAC06452.1 AB078930 BAC06454.1 AB078936 BAC06464.1 AB078935 BAC06462.1 LBMM01012027 KMQ86470.1 D85594 BAA19776.1 KZ149896 PZC78733.1 AGBW02008417 OWR53447.1 GL438017 EFN69706.1 LBMM01006671 KMQ90407.1 LBMM01008448 KMQ88867.1 LBMM01011179 KMQ86989.1 LBMM01004272 KMQ92618.1 LBMM01006954 KMQ90166.1 LBMM01007036 KMQ90099.1 LBMM01012412 KMQ86235.1 LBMM01006945 KMQ90173.1 LBMM01008278 KMQ89009.1 LBMM01006996 KMQ90130.1 LBMM01004262 KMQ92649.1 LBMM01014122 KMQ85315.1 LBMM01003941 KMQ92970.1 LBMM01009535 KMQ88053.1 LBMM01008445 KMQ88871.1 LBMM01005474 KMQ91485.1 LBMM01015285 KMQ84870.1 LBMM01003988 KMQ92922.1 LBMM01015025 KMQ84953.1 LBMM01002052 KMQ95336.1 RSAL01003504 RVE40273.1 LBMM01006977 KMQ90149.1 GGMR01015159 MBY27778.1 LBMM01004634 KMQ92224.1 GGMR01006838 MBY19457.1 LBMM01008902 KMQ88543.1 LBMM01011694 KMQ86686.1 ABLF02023257 GGMR01019493 MBY32112.1 LBMM01008422 KMQ88894.1 LBMM01006657 KMQ90417.1 ABLF02018098 ABLF02012448 ABLF02012450 ABLF02012455 ABLF02012460 LBMM01005634 KMQ91348.1 GL449531 EFN82526.1 ABLF02034925 ABLF02018808 ABLF02027248 GBHO01006398 JAG37206.1 AB593325 BAK38646.1 ABLF02019402 ABLF02059310 GBHO01002406 JAG41198.1 ABLF02011238 LBMM01016720 KMQ84328.1 GGFJ01001453 MBW50594.1 GGFJ01001454 MBW50595.1 LBMM01004367 KMQ92505.1 ABLF02014650 ABLF02030663 ABLF02041312 ABLF02018942 ABLF02041885 GFXV01002090 MBW13895.1 GDAI01000741 JAI16862.1 ABLF02009073 ABLF02030702 ABLF02042963 ABLF02011183 ABLF02041884 ABLF02007989 ABLF02004854 ABLF02041317 LBMM01007425 KMQ89740.1 ABLF02041886 LBMM01008226 KMQ89040.1 GGMR01017503 MBY30122.1 ABLF02041557 DS235827 EEB17940.1 GGFL01001916 MBW66094.1 GGMR01012748 MBY25367.1 GFXV01003903 MBW15708.1 GGFJ01001439 MBW50580.1 GGFJ01001165 MBW50306.1 GFTR01008427 JAW07999.1 GGFJ01001164 MBW50305.1 LBMM01009610 KMQ87992.1 GGFK01007771 MBW41092.1 GGFL01004044 MBW68222.1 GGFM01001733 MBW22484.1 AB090822 BAC57920.1 KU543682 AMS38369.1 GGFM01001554 MBW22305.1 GDHF01029467 GDHF01011533 JAI22847.1 JAI40781.1
Proteomes
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR002123 Plipid/glycerol_acylTrfase
IPR024079 MetalloPept_cat_dom_sf
IPR001590 Peptidase_M12B
IPR018247 EF_Hand_1_Ca_BS
IPR021896 Transposase_37
IPR012337 RNaseH-like_sf
IPR002156 RNaseH_domain
IPR036397 RNaseH_sf
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR002123 Plipid/glycerol_acylTrfase
IPR024079 MetalloPept_cat_dom_sf
IPR001590 Peptidase_M12B
IPR018247 EF_Hand_1_Ca_BS
IPR021896 Transposase_37
IPR012337 RNaseH-like_sf
IPR002156 RNaseH_domain
IPR036397 RNaseH_sf
Gene 3D
ProteinModelPortal
Q868Q4
A0A212EYI2
Q8MY29
Q8MY37
Q8MY31
Q8MY35
+ More
Q8MY33 Q8MY25 Q8MY27 A0A0J7N1D9 O01419 A0A2W1C3J5 A0A212FI97 E2AA34 A0A0J7KJG6 A0A0J7KF37 A0A0J7K9N6 A0A0J7KQK3 A0A0J7KIY5 A0A0J7KIM6 A0A0J7N0R3 A0A0J7KIC2 A0A0J7KFH2 A0A0J7NC28 A0A0J7KQY4 A0A0J7MY17 A0A0J7NKG1 A0A0J7KCE1 A0A0J7KF91 A0A0J7NFZ0 A0A0J7MWN5 A0A0J7KRB8 A0A0J7K3F1 A0A0J7KYD2 A0A3S2LNH8 A0A0J7KIS5 A0A2S2PEZ7 A0A0J7KPR0 A0A2S2NQQ6 A0A0J7KDK4 A0A0J7K903 J9KNN8 A0A2S2PRL8 A0A0J7KF40 A0A0J7KJH4 J9LA43 J9K6F0 A0A0J7KLS2 E2BP55 J9LVU7 J9KNS1 J9L0J0 A0A0A9Z010 F7IYV5 X1X2B5 A0A0A9ZCY0 X1WJY4 A0A0J7K1J9 A0A2M4BC18 A0A2M4BC24 A0A0J7NJ13 J9LM37 J9LQQ1 J9KM84 A0A2H8TIE5 A0A0K8TR60 J9KT61 X1WZ64 J9JY94 X1WXT5 A0A0J7NAV8 J9KJG8 A0A0J7N8R4 A0A2S2PKY3 J9KUM6 A0A1S4EMG7 E0VX34 A0A2M4CL98 A0A2S2P7D1 A0A2H8TR36 A0A2M4BC28 A0A2M4BB89 A0A224XIZ2 A0A2M4BB78 A0A0J7KCD9 A0A2M4AK35 A0A2M4CS98 A0A2M3Z1V8 Q868R0 A0A142LX43 A0A2M3Z1C1 A0A0K8VPH1
Q8MY33 Q8MY25 Q8MY27 A0A0J7N1D9 O01419 A0A2W1C3J5 A0A212FI97 E2AA34 A0A0J7KJG6 A0A0J7KF37 A0A0J7K9N6 A0A0J7KQK3 A0A0J7KIY5 A0A0J7KIM6 A0A0J7N0R3 A0A0J7KIC2 A0A0J7KFH2 A0A0J7NC28 A0A0J7KQY4 A0A0J7MY17 A0A0J7NKG1 A0A0J7KCE1 A0A0J7KF91 A0A0J7NFZ0 A0A0J7MWN5 A0A0J7KRB8 A0A0J7K3F1 A0A0J7KYD2 A0A3S2LNH8 A0A0J7KIS5 A0A2S2PEZ7 A0A0J7KPR0 A0A2S2NQQ6 A0A0J7KDK4 A0A0J7K903 J9KNN8 A0A2S2PRL8 A0A0J7KF40 A0A0J7KJH4 J9LA43 J9K6F0 A0A0J7KLS2 E2BP55 J9LVU7 J9KNS1 J9L0J0 A0A0A9Z010 F7IYV5 X1X2B5 A0A0A9ZCY0 X1WJY4 A0A0J7K1J9 A0A2M4BC18 A0A2M4BC24 A0A0J7NJ13 J9LM37 J9LQQ1 J9KM84 A0A2H8TIE5 A0A0K8TR60 J9KT61 X1WZ64 J9JY94 X1WXT5 A0A0J7NAV8 J9KJG8 A0A0J7N8R4 A0A2S2PKY3 J9KUM6 A0A1S4EMG7 E0VX34 A0A2M4CL98 A0A2S2P7D1 A0A2H8TR36 A0A2M4BC28 A0A2M4BB89 A0A224XIZ2 A0A2M4BB78 A0A0J7KCD9 A0A2M4AK35 A0A2M4CS98 A0A2M3Z1V8 Q868R0 A0A142LX43 A0A2M3Z1C1 A0A0K8VPH1
PDB
1WDU
E-value=2.60887e-11,
Score=167
Ontologies
GO
Topology
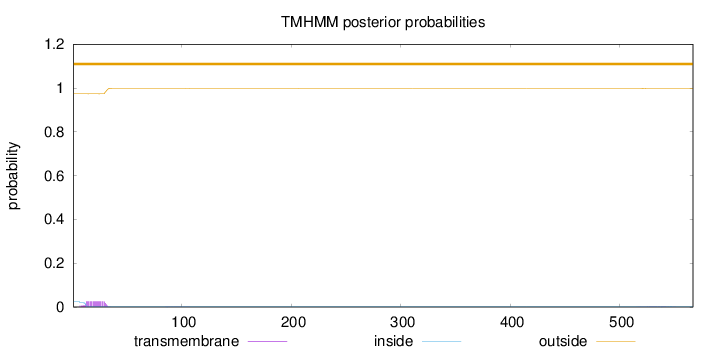
Length:
567
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.568489999999999
Exp number, first 60 AAs:
0.51711
Total prob of N-in:
0.02597
outside
1 - 567
Population Genetic Test Statistics
Pi
0.007866
Theta
0.056452
Tajima's D
-0.504031
CLR
43.034583
CSRT
0.088045597720114
Interpretation
Uncertain
