Gene
KWMTBOMO07356 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010679
Annotation
glutamyl_aminopeptidase-like_[Bombyx_mori]
Full name
Aminopeptidase
Location in the cell
PlasmaMembrane Reliability : 1.569
Sequence
CDS
ATGTCTCCGAAACTCTCACAATGCTTATTTAGAAACAACATCATCATTCTTCTAGGGCTTTTGTCGTTTTCCTTAATAGCCCCAGCAATTACGTTGACGACTTTTGACAAAAGGTCCAAAACATGTGAAAGCGATGACTGCGTGCCCATGGCTGTCAGAGAAGAAACGAATTTGTCAAACGTAACTGAGAATGCAAATTACTACAGACTGCCTAACAACATCCTCCCTAAAAACTATAATCTGACATTGAAGCCGAACATGAAAGATAATATTTTCAAAGGCACAGTCAACATTGCAATAGAAGTACTGACATCTACGAATAAAATCACCATGCATGCTTATAAGTTAACTATTGAATCCGTGATTTTGAAAGATTCGAAGAACGTGGAAGTCTCAATATCAAGTATAAACATAAGTACCGATAAAAGAGAATTGCTCAGAATACACTTAAATGATCAAATACAGAGGGGAAAATACAATGTGGAAATTGTATTCCAAGGAAACATGGATAAGAAAATAATTGGGTTTTACTCAAGTTCTTTGAAAAACGGAGGAACAATGGTAGCCAGTAAATTTCAGCCAACATATGCTCGCCAAGCATTCCCTTGTTTTGATGAGCCAGATTTCAAAGCCACATATGATATTGCACTTGTTAAACCCGAAGGGTATGTGGCATTGTCAAACATGAATGAAATATCAGTGACACGGGACCCATCATCTGATTTAGAAACTGTCAAATTTGCGACCAGTGTCCCAATGTCGACATATCTGGCTTGTTTTGTTGTATGTGATTTTGGATATAAGGATGTTGAAATTAATACTTCTGGTATCGGAAATACTTTCAAACTTAGATCTTTTGCTCAAAAAAATGAACTTCATAAGATTGATTTTGCTCAAGACATTGGGAAGAGAGCTACTGAATTTTACATAAGATATTATGAAGTAGAATTTCCCTTGCCAAAATTAGATATGATTGCAATACCTGACTACATTTCTGGAGCCACTGAGCACTGGGGTCTTATAACATACAGAGAAACATCATTTCTGGTTGATGAGGCTACTGCTTCTGTTAAAAATAAGATCAGTATTGCTAATACTATAGCTCATGAACTGGCTCACATGTGGTTTGGAAATTTAGTGACAATGAAATGGTGGGATGAAGTATGGCTAAATGAAGGTTTTGCTTCCTACATGCAAGTTAAATCATTGAATGCAATTGAACCATCGTGGGCCATGTTGGATCAATTCTTAACAAAAACTGTTCATCCTGTACTAGTGACTGATGCCAAACTTTCAAGTCATCCTATAGTACAAACTGTGTCTACACCCGATCAGATAACATCAATATTTGACACAATTTCTTACAATAAAGGCGCATCCATATTAAGAATGCTCGAGGGTTTCATTGGCGAAGAAAACTTCCGTCGTGGCGTTTCAGATTACCTCAAGAAGTTTCAGTACGGTAATACAGTGACTCAAGATTTACTATCCTGTCTTGAAGTTTACTTTAAACAGGAAAATCCTGATTTAAGCCTGACTCACATCATGGATACGTGGACACAACAAATGGGTTATCCACTACTCTACGTTGAACCGGGGAATGGAACAAACACATACGTTGTTACTCAAAAACGGTTTTTGCTCGATCCTGACGCAGAATACACGAACGATTCGAAGTTCAATTACCGTTGGTACGTGCCGATAACTTACAAGACAAATAAAGGAAATTGCGGCAGAGTTATTTGGTTCCCCGATACGGTAGAGAGCGTAACACTGAATCTCGATGACAATATCAGATGGCTGAAAATTAATAACAATCAAATTGGTTATTATAGAGTTCATTATGCCGATGAGATGTGGCAAAATCTTATACTAGAGTTAAACGCTAAATCGAAAGAGTTAACAATATCAGATCGAGCGCATTTATTGGATGACGCGTTTGCTCTCGCCGAAGCCGGCTCGTTGCCTTACAATATAGCGTTGGATCTCACAACGTATCTCACTGTGGAGGATGATGAAATACCTTGGACGACCGCCGTATCAATATTCGGAGGCTTAGCCAGAAGATTGCTGAACACACCAGCGTACGATGACCTTAAGAGCTATATCCATGGTCTAGTGAAACCTGTATACGAGAAACAAAGTTGGGAAAAGGTTAATATTGGAGTTATTGAAAGGCTACTTCGAGCAAGGATACTAACTCTAGCAACTCGTTACCAATTGCCAGACGCCGAAGATAAAGTAAGAAATTTGTTCTTGAGTTGGCTGAACGGTCACGGTACTCCCGATGCCGTTGTGATCGAACCAGATTTGCGGGACTTTGTTTATTATTATGGAATGAGATCGGCCACTCAGCAAGAATGGGATAAGTTGTGGGAGATCTATTTAAAAGAGACGGACGTTCTCGAAGCCACGAAAATTCGAAGCGCCCTATCGGCGTCTCGTGACGCGAACATCTTAAAAAGATACTTGGAGCTGTCTTGGGACGAAGCGAACATTCGGAGCCAGGATTACTTAAACGTTTTAGCGGACGTCAGCGACAACCCTACCGGCACCGGTCTGGTTTGGGACGACGTGAGGACGCGATGGCCCCAGCTAGTCGACCGATTCACGTTGAACAGCCGTTACTTGGGCGGCCTCATTACCAGCATCACGAAATCATTTAGCACACAGCAAAAACTAAACGAGATGGAAGCATTCTTCGCTCAATATCCAGAAGCTGGTGCTGGCGAGGCCTCAAGAAAACGTGCTCTAGAAACAGTTCACAACAACATCAAGTGGTCTCAGAAGCACCGAGCGTCTGTCGCAGCCTGGCTGGAGAAACGTCGTTCTTGA
Protein
MSPKLSQCLFRNNIIILLGLLSFSLIAPAITLTTFDKRSKTCESDDCVPMAVREETNLSNVTENANYYRLPNNILPKNYNLTLKPNMKDNIFKGTVNIAIEVLTSTNKITMHAYKLTIESVILKDSKNVEVSISSINISTDKRELLRIHLNDQIQRGKYNVEIVFQGNMDKKIIGFYSSSLKNGGTMVASKFQPTYARQAFPCFDEPDFKATYDIALVKPEGYVALSNMNEISVTRDPSSDLETVKFATSVPMSTYLACFVVCDFGYKDVEINTSGIGNTFKLRSFAQKNELHKIDFAQDIGKRATEFYIRYYEVEFPLPKLDMIAIPDYISGATEHWGLITYRETSFLVDEATASVKNKISIANTIAHELAHMWFGNLVTMKWWDEVWLNEGFASYMQVKSLNAIEPSWAMLDQFLTKTVHPVLVTDAKLSSHPIVQTVSTPDQITSIFDTISYNKGASILRMLEGFIGEENFRRGVSDYLKKFQYGNTVTQDLLSCLEVYFKQENPDLSLTHIMDTWTQQMGYPLLYVEPGNGTNTYVVTQKRFLLDPDAEYTNDSKFNYRWYVPITYKTNKGNCGRVIWFPDTVESVTLNLDDNIRWLKINNNQIGYYRVHYADEMWQNLILELNAKSKELTISDRAHLLDDAFALAEAGSLPYNIALDLTTYLTVEDDEIPWTTAVSIFGGLARRLLNTPAYDDLKSYIHGLVKPVYEKQSWEKVNIGVIERLLRARILTLATRYQLPDAEDKVRNLFLSWLNGHGTPDAVVIEPDLRDFVYYYGMRSATQQEWDKLWEIYLKETDVLEATKIRSALSASRDANILKRYLELSWDEANIRSQDYLNVLADVSDNPTGTGLVWDDVRTRWPQLVDRFTLNSRYLGGLITSITKSFSTQQKLNEMEAFFAQYPEAGAGEASRKRALETVHNNIKWSQKHRASVAAWLEKRRS
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M1 family.
Feature
chain Aminopeptidase
Uniprot
I3VR80
H9JMC6
A0A194PNI2
A0A3S2LS36
A0A2A4JGY4
A0A286LPZ9
+ More
A0A1L8EJF3 A0A1L8EJ79 A0A1L8EJH8 A0A1I8MDK7 A0A1I8PTD3 A0A1W4U7A0 W8AS98 A0A0L0CEH0 B3P162 A0A0K8V4C0 B4M6C7 B4JEL8 B4G3L4 A0A0Q9X6W1 A0A0A1WLH6 Q9VFX0 Q29A14 Q86NQ5 A0A0M3QXN4 A0A0K8TTQ5 B4HG90 A0A1I8PGY6 A0A3B0K3M7 A0A0A1WTT6 A0A336LYS0 A0A1I8MQF9 A0A2J7PNK0 T1PDR7 A0A0L0CE23 A0A1L8DP89 A0A1A9VKN8 A0A1I8MQG5 A0A1L8EJB5 A0A067REW1 A0A1L8EJ70 A0A034VI37 A0A1Y1KMG6 A0A0K8V2U8 A0A0A1WY54 A0A1Y1KJS9 A0A0K8UPF8 A0A034VBA9 A0A1A9Y906 A0A1A9VYF7 A0A1B0BGT8 U5EU90 E2C1U5 B4G3L5 A0A1I8P3V9 W8AYS9 A0A1Y1KMH6 A0A1I8P3R0 A0A1I8NG19 A0A1I8Q3H2 A0A1B0AGP0 A0A182K7Z1 A0A1B0G7K8 A0A034VKU0 Q8IHC5 A0A034VK36 B4HG89 A0A1W4WP24 Q9VFW9 A9UNG9 Q9VFW8 A0A0K8US39 B4R1B3 A0A0Q9WUY2 B3P161 A0A1W4WCU2 A0A1B6C9L0 F5HKY0 A0A182HIX8 A0A182P2X3 A0A182X8C3 A0A182G9S4 A0A3B0K5B1 A0A0P6IZ91 A0A182XVQ3 A0A182USH2 A0A1B6EQM6 A0A182JEQ2 A0A1B6FX35 A0A182TTF3 A0A023EY50 A0A1Q3EZL5 Q173A7 A0A0P6J4X5 J9K7S8 A0A2A3E257 A0A1L8E630
A0A1L8EJF3 A0A1L8EJ79 A0A1L8EJH8 A0A1I8MDK7 A0A1I8PTD3 A0A1W4U7A0 W8AS98 A0A0L0CEH0 B3P162 A0A0K8V4C0 B4M6C7 B4JEL8 B4G3L4 A0A0Q9X6W1 A0A0A1WLH6 Q9VFX0 Q29A14 Q86NQ5 A0A0M3QXN4 A0A0K8TTQ5 B4HG90 A0A1I8PGY6 A0A3B0K3M7 A0A0A1WTT6 A0A336LYS0 A0A1I8MQF9 A0A2J7PNK0 T1PDR7 A0A0L0CE23 A0A1L8DP89 A0A1A9VKN8 A0A1I8MQG5 A0A1L8EJB5 A0A067REW1 A0A1L8EJ70 A0A034VI37 A0A1Y1KMG6 A0A0K8V2U8 A0A0A1WY54 A0A1Y1KJS9 A0A0K8UPF8 A0A034VBA9 A0A1A9Y906 A0A1A9VYF7 A0A1B0BGT8 U5EU90 E2C1U5 B4G3L5 A0A1I8P3V9 W8AYS9 A0A1Y1KMH6 A0A1I8P3R0 A0A1I8NG19 A0A1I8Q3H2 A0A1B0AGP0 A0A182K7Z1 A0A1B0G7K8 A0A034VKU0 Q8IHC5 A0A034VK36 B4HG89 A0A1W4WP24 Q9VFW9 A9UNG9 Q9VFW8 A0A0K8US39 B4R1B3 A0A0Q9WUY2 B3P161 A0A1W4WCU2 A0A1B6C9L0 F5HKY0 A0A182HIX8 A0A182P2X3 A0A182X8C3 A0A182G9S4 A0A3B0K5B1 A0A0P6IZ91 A0A182XVQ3 A0A182USH2 A0A1B6EQM6 A0A182JEQ2 A0A1B6FX35 A0A182TTF3 A0A023EY50 A0A1Q3EZL5 Q173A7 A0A0P6J4X5 J9K7S8 A0A2A3E257 A0A1L8E630
EC Number
3.4.11.-
Pubmed
EMBL
JQ061151
AFK85025.1
BABH01033483
BABH01033484
KQ459603
KPI92680.1
+ More
RSAL01000020 RVE52664.1 NWSH01001630 PCG70652.1 MF425656 ASU92548.1 GFDG01000011 JAV18788.1 GFDG01000009 JAV18790.1 GFDG01000010 JAV18789.1 GAMC01017598 JAB88957.1 JRES01000501 KNC30646.1 CH954181 EDV49251.1 GDHF01032702 GDHF01018595 JAI19612.1 JAI33719.1 CH940652 EDW59203.2 CH916369 EDV93149.1 CH479179 EDW25022.1 CH933806 KRG01076.1 GBXI01014942 JAC99349.1 AE014297 BT133114 AAF54925.2 AEY64282.1 CM000070 EAL27537.2 BT003783 AAO41464.1 CP012526 ALC46233.1 GDAI01000077 JAI17526.1 CH480815 EDW42338.1 OUUW01000013 SPP87883.1 GBXI01012226 JAD02066.1 UFQT01000315 SSX23132.1 NEVH01023953 PNF17923.1 KA646839 AFP61468.1 KNC30663.1 GFDF01005802 JAV08282.1 GFDG01000018 JAV18781.1 KK852680 KDR18594.1 GFDG01000019 JAV18780.1 GAKP01016801 GAKP01016800 GAKP01016798 JAC42154.1 GEZM01083993 JAV60865.1 GDHF01019399 JAI32915.1 GBXI01010989 JAD03303.1 GEZM01083994 GEZM01083991 JAV60868.1 GDHF01023725 JAI28589.1 GAKP01019168 JAC39784.1 JXJN01013993 GANO01002425 JAB57446.1 GL452008 EFN78057.1 EDW25023.1 GAMC01016533 GAMC01016532 JAB90023.1 GEZM01083987 JAV60875.1 CCAG010018818 CCAG010018819 CCAG010018820 GAKP01016799 JAC42153.1 BT001313 AAN71068.1 GAKP01016802 JAC42150.1 EDW42337.1 AAF54926.3 BT031333 ABY21746.1 AAF54927.2 GDHF01023154 JAI29160.1 CM000364 EDX13090.1 CH964272 KRF99951.1 EDV49250.1 GEDC01027157 JAS10141.1 AAAB01008879 EGK96941.1 APCN01002103 APCN01002104 JXUM01049783 JXUM01049784 JXUM01049785 KQ561620 KXJ77985.1 SPP87882.1 GDUN01000766 JAN95153.1 GECZ01029696 JAS40073.1 AXCP01008598 GECZ01015088 JAS54681.1 GAPW01000124 JAC13474.1 GFDL01014303 JAV20742.1 CH477428 EAT41121.1 GDUN01000765 JAN95154.1 ABLF02038188 KZ288431 PBC25823.1 GFDG01004631 JAV14168.1
RSAL01000020 RVE52664.1 NWSH01001630 PCG70652.1 MF425656 ASU92548.1 GFDG01000011 JAV18788.1 GFDG01000009 JAV18790.1 GFDG01000010 JAV18789.1 GAMC01017598 JAB88957.1 JRES01000501 KNC30646.1 CH954181 EDV49251.1 GDHF01032702 GDHF01018595 JAI19612.1 JAI33719.1 CH940652 EDW59203.2 CH916369 EDV93149.1 CH479179 EDW25022.1 CH933806 KRG01076.1 GBXI01014942 JAC99349.1 AE014297 BT133114 AAF54925.2 AEY64282.1 CM000070 EAL27537.2 BT003783 AAO41464.1 CP012526 ALC46233.1 GDAI01000077 JAI17526.1 CH480815 EDW42338.1 OUUW01000013 SPP87883.1 GBXI01012226 JAD02066.1 UFQT01000315 SSX23132.1 NEVH01023953 PNF17923.1 KA646839 AFP61468.1 KNC30663.1 GFDF01005802 JAV08282.1 GFDG01000018 JAV18781.1 KK852680 KDR18594.1 GFDG01000019 JAV18780.1 GAKP01016801 GAKP01016800 GAKP01016798 JAC42154.1 GEZM01083993 JAV60865.1 GDHF01019399 JAI32915.1 GBXI01010989 JAD03303.1 GEZM01083994 GEZM01083991 JAV60868.1 GDHF01023725 JAI28589.1 GAKP01019168 JAC39784.1 JXJN01013993 GANO01002425 JAB57446.1 GL452008 EFN78057.1 EDW25023.1 GAMC01016533 GAMC01016532 JAB90023.1 GEZM01083987 JAV60875.1 CCAG010018818 CCAG010018819 CCAG010018820 GAKP01016799 JAC42153.1 BT001313 AAN71068.1 GAKP01016802 JAC42150.1 EDW42337.1 AAF54926.3 BT031333 ABY21746.1 AAF54927.2 GDHF01023154 JAI29160.1 CM000364 EDX13090.1 CH964272 KRF99951.1 EDV49250.1 GEDC01027157 JAS10141.1 AAAB01008879 EGK96941.1 APCN01002103 APCN01002104 JXUM01049783 JXUM01049784 JXUM01049785 KQ561620 KXJ77985.1 SPP87882.1 GDUN01000766 JAN95153.1 GECZ01029696 JAS40073.1 AXCP01008598 GECZ01015088 JAS54681.1 GAPW01000124 JAC13474.1 GFDL01014303 JAV20742.1 CH477428 EAT41121.1 GDUN01000765 JAN95154.1 ABLF02038188 KZ288431 PBC25823.1 GFDG01004631 JAV14168.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000218220
UP000095301
UP000095300
+ More
UP000192221 UP000037069 UP000008711 UP000008792 UP000001070 UP000008744 UP000009192 UP000000803 UP000001819 UP000092553 UP000001292 UP000268350 UP000235965 UP000078200 UP000027135 UP000092443 UP000092460 UP000008237 UP000092445 UP000075881 UP000092444 UP000192223 UP000000304 UP000007798 UP000007062 UP000075840 UP000075885 UP000076407 UP000069940 UP000249989 UP000076408 UP000075903 UP000075880 UP000075902 UP000008820 UP000007819 UP000242457
UP000192221 UP000037069 UP000008711 UP000008792 UP000001070 UP000008744 UP000009192 UP000000803 UP000001819 UP000092553 UP000001292 UP000268350 UP000235965 UP000078200 UP000027135 UP000092443 UP000092460 UP000008237 UP000092445 UP000075881 UP000092444 UP000192223 UP000000304 UP000007798 UP000007062 UP000075840 UP000075885 UP000076407 UP000069940 UP000249989 UP000076408 UP000075903 UP000075880 UP000075902 UP000008820 UP000007819 UP000242457
Interpro
Gene 3D
CDD
ProteinModelPortal
I3VR80
H9JMC6
A0A194PNI2
A0A3S2LS36
A0A2A4JGY4
A0A286LPZ9
+ More
A0A1L8EJF3 A0A1L8EJ79 A0A1L8EJH8 A0A1I8MDK7 A0A1I8PTD3 A0A1W4U7A0 W8AS98 A0A0L0CEH0 B3P162 A0A0K8V4C0 B4M6C7 B4JEL8 B4G3L4 A0A0Q9X6W1 A0A0A1WLH6 Q9VFX0 Q29A14 Q86NQ5 A0A0M3QXN4 A0A0K8TTQ5 B4HG90 A0A1I8PGY6 A0A3B0K3M7 A0A0A1WTT6 A0A336LYS0 A0A1I8MQF9 A0A2J7PNK0 T1PDR7 A0A0L0CE23 A0A1L8DP89 A0A1A9VKN8 A0A1I8MQG5 A0A1L8EJB5 A0A067REW1 A0A1L8EJ70 A0A034VI37 A0A1Y1KMG6 A0A0K8V2U8 A0A0A1WY54 A0A1Y1KJS9 A0A0K8UPF8 A0A034VBA9 A0A1A9Y906 A0A1A9VYF7 A0A1B0BGT8 U5EU90 E2C1U5 B4G3L5 A0A1I8P3V9 W8AYS9 A0A1Y1KMH6 A0A1I8P3R0 A0A1I8NG19 A0A1I8Q3H2 A0A1B0AGP0 A0A182K7Z1 A0A1B0G7K8 A0A034VKU0 Q8IHC5 A0A034VK36 B4HG89 A0A1W4WP24 Q9VFW9 A9UNG9 Q9VFW8 A0A0K8US39 B4R1B3 A0A0Q9WUY2 B3P161 A0A1W4WCU2 A0A1B6C9L0 F5HKY0 A0A182HIX8 A0A182P2X3 A0A182X8C3 A0A182G9S4 A0A3B0K5B1 A0A0P6IZ91 A0A182XVQ3 A0A182USH2 A0A1B6EQM6 A0A182JEQ2 A0A1B6FX35 A0A182TTF3 A0A023EY50 A0A1Q3EZL5 Q173A7 A0A0P6J4X5 J9K7S8 A0A2A3E257 A0A1L8E630
A0A1L8EJF3 A0A1L8EJ79 A0A1L8EJH8 A0A1I8MDK7 A0A1I8PTD3 A0A1W4U7A0 W8AS98 A0A0L0CEH0 B3P162 A0A0K8V4C0 B4M6C7 B4JEL8 B4G3L4 A0A0Q9X6W1 A0A0A1WLH6 Q9VFX0 Q29A14 Q86NQ5 A0A0M3QXN4 A0A0K8TTQ5 B4HG90 A0A1I8PGY6 A0A3B0K3M7 A0A0A1WTT6 A0A336LYS0 A0A1I8MQF9 A0A2J7PNK0 T1PDR7 A0A0L0CE23 A0A1L8DP89 A0A1A9VKN8 A0A1I8MQG5 A0A1L8EJB5 A0A067REW1 A0A1L8EJ70 A0A034VI37 A0A1Y1KMG6 A0A0K8V2U8 A0A0A1WY54 A0A1Y1KJS9 A0A0K8UPF8 A0A034VBA9 A0A1A9Y906 A0A1A9VYF7 A0A1B0BGT8 U5EU90 E2C1U5 B4G3L5 A0A1I8P3V9 W8AYS9 A0A1Y1KMH6 A0A1I8P3R0 A0A1I8NG19 A0A1I8Q3H2 A0A1B0AGP0 A0A182K7Z1 A0A1B0G7K8 A0A034VKU0 Q8IHC5 A0A034VK36 B4HG89 A0A1W4WP24 Q9VFW9 A9UNG9 Q9VFW8 A0A0K8US39 B4R1B3 A0A0Q9WUY2 B3P161 A0A1W4WCU2 A0A1B6C9L0 F5HKY0 A0A182HIX8 A0A182P2X3 A0A182X8C3 A0A182G9S4 A0A3B0K5B1 A0A0P6IZ91 A0A182XVQ3 A0A182USH2 A0A1B6EQM6 A0A182JEQ2 A0A1B6FX35 A0A182TTF3 A0A023EY50 A0A1Q3EZL5 Q173A7 A0A0P6J4X5 J9K7S8 A0A2A3E257 A0A1L8E630
PDB
4KXD
E-value=5.63718e-158,
Score=1434
Ontologies
GO
Topology
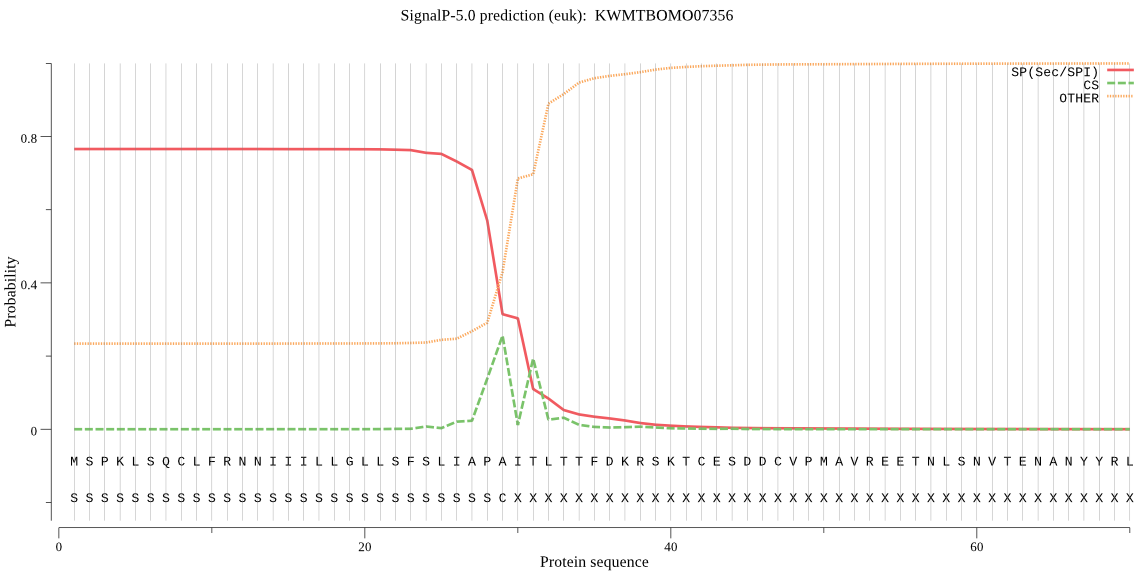
SignalP
Position: 1 - 29,
Likelihood: 0.766682
Length:
944
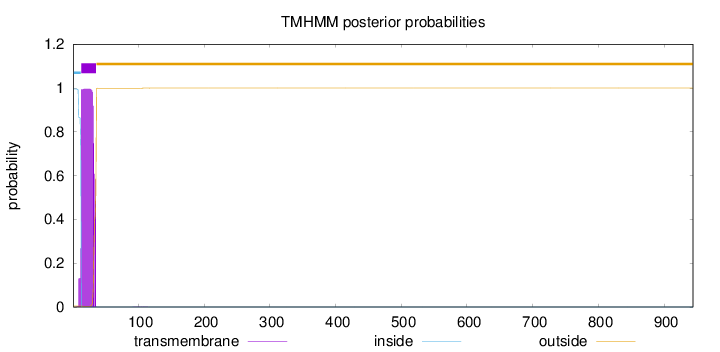
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.9171999999999
Exp number, first 60 AAs:
21.9081
Total prob of N-in:
0.99571
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 944
Population Genetic Test Statistics
Pi
20.5103
Theta
19.856299
Tajima's D
0.249694
CLR
0.824573
CSRT
0.456627168641568
Interpretation
Uncertain
