Pre Gene Modal
BGIBMGA010676
Annotation
PREDICTED:_stearoyl-CoA_desaturase_5_isoform_X2_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.604
Sequence
CDS
ATGGCCTTCCAAAATCATATATATGAATGGGTACGCGATCATAGAGTTCATCATAAGTTCACGGAGACTGACGCTGACCCTCACAACGCTAAACGAGGATTTTTCTTTTCTCACATGGGCTGGTTGATGGTGAGGAAGCACAAAGACGTCTTCGAAAAGGGTGCCGCTGTTGATATGTCGGATTTGGAAAAGGACCCGATCGTAATGTTCCAAAAAAAAACATATCTATTCGTGATGCCAATCCTATGCTTCTTATTGCCATTGTACATTCCCGTTTATTTTTGGAACGAAAATCCATGGTCCTCGTGGTATGTCGCATCAATGTTTAGATACACGATCAGTCTTCACTTCACGTGGCTGGTTAACTCCGCCGCACACCTCTGGGGGAACAAGCCATATGATCAATACATAGGAGCAACTGATAACAAAACTGTGGCGATCTGTGCTTTAGGCGAGGGTTGGCACAACTATCATCACGTATTTCCCTGGGACTACAAAGCGGCCGAGCTCGGAGACTACAGCACCAATTTATCAACCGCCTTAATTGACATCGCTGCAAAATACGGCTTAGCTTACGATTTGAAGACGGTATCTGAAAAGATGATAAGGAATCGCATTAATCGAACAGGCGACGGTACCCATCCTTGGGCGAAACAGAAGGCAGAGTTGGAGGAAGATCATCATCATCTAGAAAACCCTATATGGGGCTGGGGCGATAAAGACATGTCAGACGATGATAAGAAATTGGCGGAAATTGTTCACAAAAAGAATGACTAG
Protein
MAFQNHIYEWVRDHRVHHKFTETDADPHNAKRGFFFSHMGWLMVRKHKDVFEKGAAVDMSDLEKDPIVMFQKKTYLFVMPILCFLLPLYIPVYFWNENPWSSWYVASMFRYTISLHFTWLVNSAAHLWGNKPYDQYIGATDNKTVAICALGEGWHNYHHVFPWDYKAAELGDYSTNLSTALIDIAAKYGLAYDLKTVSEKMIRNRINRTGDGTHPWAKQKAELEEDHHHLENPIWGWGDKDMSDDDKKLAEIVHKKND
Summary
Cofactor
Fe cation
Similarity
Belongs to the fatty acid desaturase type 1 family.
Uniprot
H9JMC3
A0A212F2M1
A0A194R480
A0A068FRE6
A0A194PH02
B7SB79
+ More
Q8MZZ5 A0A0U2F3D3 A0A0U2F3C0 I7AHL6 A0A2H1VBX7 A0A2A4JFV9 T1RTG7 A0A1L8D6C3 T1RTD2 S4PN29 A0A0M3LAH8 B6CBS4 A0A0L7LIJ1 Q6A4M8 I4DS07 D8UU48 A0A2P8ZGJ8 A0A1B0DIJ1 A0A2J7PNJ8 A0A151JS75 A0A154PDD1 A0A1B6BZA0 K7J0U9 A0A232F3U6 A0A1B6DXZ2 Q4PP96 E2C1U4 A0A0J7NME9 A0A2J7Q458 A0A1S4EHT5 A0A2P8XTS3 E2AX12 F4W5A9 A0A0C9QWJ2 T1HR83 Q173B1 A0A0V0GDS0 A0A0P6J0V5 A0A195FTD4 A0A067QT59 A0A1W4X6D0 A0A310SQM5 A0A151X2W9 A0A195CXM1 Q5XXT0 A0A1S4EFC2 A0A1S3D6B4 A0A069DSW7 A0A023ERW8 E0VE45 A0A1S3D7T3 A0A026W4L1 Q9BH41 A0A1B6GKP5 A0A182TJZ6 A0A195BTK0 A0A1L2BNU6 Q6PPH9 A0A023ERW2 A0A182LZE7 A0A182K1K4 D7EJN1 A0A096XDC8 J9K131 A0A0L7RGE5 A0A182KWY6 A0A2H8TMD9 T1RLC5 A0A158NYT0 A0A182HJS3 A0A182XK65 A0A067RHH4 A0A182T7Z9 A0A182V4X1 A0A1B6ER20 T1RLF5 Q201V1 E0VE42 Q7QDJ5 A0A2S2PCR7 A0A3Q0JL17 A0A182PS55 A0A182W0K0 A0A2R7W9M0 A0A2A3E2X3 V9IBM6 A0A182Y4H9 A0A182QWH3 A0A2S2PLC4 A0A088AI91 A0A182R814 A0A182IPM3 W5JSC3 A0A1I8QDS2
Q8MZZ5 A0A0U2F3D3 A0A0U2F3C0 I7AHL6 A0A2H1VBX7 A0A2A4JFV9 T1RTG7 A0A1L8D6C3 T1RTD2 S4PN29 A0A0M3LAH8 B6CBS4 A0A0L7LIJ1 Q6A4M8 I4DS07 D8UU48 A0A2P8ZGJ8 A0A1B0DIJ1 A0A2J7PNJ8 A0A151JS75 A0A154PDD1 A0A1B6BZA0 K7J0U9 A0A232F3U6 A0A1B6DXZ2 Q4PP96 E2C1U4 A0A0J7NME9 A0A2J7Q458 A0A1S4EHT5 A0A2P8XTS3 E2AX12 F4W5A9 A0A0C9QWJ2 T1HR83 Q173B1 A0A0V0GDS0 A0A0P6J0V5 A0A195FTD4 A0A067QT59 A0A1W4X6D0 A0A310SQM5 A0A151X2W9 A0A195CXM1 Q5XXT0 A0A1S4EFC2 A0A1S3D6B4 A0A069DSW7 A0A023ERW8 E0VE45 A0A1S3D7T3 A0A026W4L1 Q9BH41 A0A1B6GKP5 A0A182TJZ6 A0A195BTK0 A0A1L2BNU6 Q6PPH9 A0A023ERW2 A0A182LZE7 A0A182K1K4 D7EJN1 A0A096XDC8 J9K131 A0A0L7RGE5 A0A182KWY6 A0A2H8TMD9 T1RLC5 A0A158NYT0 A0A182HJS3 A0A182XK65 A0A067RHH4 A0A182T7Z9 A0A182V4X1 A0A1B6ER20 T1RLF5 Q201V1 E0VE42 Q7QDJ5 A0A2S2PCR7 A0A3Q0JL17 A0A182PS55 A0A182W0K0 A0A2R7W9M0 A0A2A3E2X3 V9IBM6 A0A182Y4H9 A0A182QWH3 A0A2S2PLC4 A0A088AI91 A0A182R814 A0A182IPM3 W5JSC3 A0A1I8QDS2
Pubmed
19121390
22118469
26354079
26385554
20403437
12524345
+ More
12770579 28986331 24500170 23622113 18831750 26227816 15527989 22651552 29403074 20075255 28648823 20798317 21719571 17510324 26999592 24845553 26334808 24945155 20566863 24508170 30249741 12429125 18362917 19820115 24880119 20966253 23727612 21347285 12364791 14747013 17210077 25244985 20920257 23761445
12770579 28986331 24500170 23622113 18831750 26227816 15527989 22651552 29403074 20075255 28648823 20798317 21719571 17510324 26999592 24845553 26334808 24945155 20566863 24508170 30249741 12429125 18362917 19820115 24880119 20966253 23727612 21347285 12364791 14747013 17210077 25244985 20920257 23761445
EMBL
BABH01033480
AGBW02010723
OWR47976.1
KQ460845
KPJ12045.1
KJ622057
+ More
AID66665.1 KQ459603 KPI92676.1 EU152405 ABX71814.1 AF482905 AAM28480.2 KP008116 MF687524 AKU76403.1 ATJ44450.1 KP008123 MF687584 AKU76410.1 ATJ44510.1 JX126484 AFO38465.1 ODYU01001713 SOQ38301.1 NWSH01001630 PCG70656.1 JX531661 AGO45846.1 GEYN01000131 JAV01998.1 JX531655 AGO45840.1 GAIX01003235 JAA89325.1 KM366792 AIM40222.1 EU152334 ABX71629.1 JTDY01000964 KOB75262.1 AF518008 AF518009 AAQ12887.1 AAQ12888.1 AK405358 BAM20697.1 GQ130306 ADC53487.1 PYGN01000064 PSN55622.1 AJVK01062643 NEVH01023953 PNF17920.1 KQ978543 KYN30207.1 KQ434878 KZC09843.1 GEDC01030710 JAS06588.1 NNAY01001086 OXU25189.1 GEDC01006752 JAS30546.1 DQ062217 AAY63972.1 GL452008 EFN78056.1 LBMM01003340 KMQ93645.1 NEVH01018390 PNF23355.1 PYGN01001363 PSN35395.1 GL443495 EFN62032.1 GL887645 EGI70559.1 GBYB01008109 JAG77876.1 ACPB03003193 CH477428 EAT41117.1 GECL01000276 JAP05848.1 GDUN01000309 JAN95610.1 KQ981276 KYN43681.1 KK853306 KDR08705.1 KQ761655 OAD57203.1 KQ982566 KYQ54761.1 KQ977220 KYN04894.1 AY725782 AAU95195.1 GBGD01001849 JAC87040.1 GAPW01002159 JAC11439.1 DS235088 EEB11651.1 KK107419 QOIP01000003 EZA51020.1 RLU25069.1 AF338465 AF338466 AAK25796.1 AAK25797.1 GECZ01006781 JAS62988.1 KQ976407 KYM91399.1 KU296958 ALT55114.1 AY588067 AAT01079.1 GAPW01001631 JAC11967.1 AXCM01000236 KQ971422 EFA12798.2 KF576670 AHH30811.1 ABLF02029597 KQ414596 KOC69925.1 GFXV01003521 MBW15326.1 KC437331 AGK82081.1 ADTU01004257 APCN01002208 KK852680 KDR18593.1 GECZ01029586 JAS40183.1 KC437330 AGK82080.1 ABLF02009131 DQ413235 AK341785 ABD72703.1 BAH72082.1 EEB11648.1 AAAB01008851 EAA07365.5 GGMR01014628 MBY27247.1 KK854500 PTY16384.1 KZ288431 PBC25824.1 JR037914 AEY58032.1 AXCN02000306 AXCN02000307 GGMR01017596 MBY30215.1 ADMH02000313 ETN67016.1
AID66665.1 KQ459603 KPI92676.1 EU152405 ABX71814.1 AF482905 AAM28480.2 KP008116 MF687524 AKU76403.1 ATJ44450.1 KP008123 MF687584 AKU76410.1 ATJ44510.1 JX126484 AFO38465.1 ODYU01001713 SOQ38301.1 NWSH01001630 PCG70656.1 JX531661 AGO45846.1 GEYN01000131 JAV01998.1 JX531655 AGO45840.1 GAIX01003235 JAA89325.1 KM366792 AIM40222.1 EU152334 ABX71629.1 JTDY01000964 KOB75262.1 AF518008 AF518009 AAQ12887.1 AAQ12888.1 AK405358 BAM20697.1 GQ130306 ADC53487.1 PYGN01000064 PSN55622.1 AJVK01062643 NEVH01023953 PNF17920.1 KQ978543 KYN30207.1 KQ434878 KZC09843.1 GEDC01030710 JAS06588.1 NNAY01001086 OXU25189.1 GEDC01006752 JAS30546.1 DQ062217 AAY63972.1 GL452008 EFN78056.1 LBMM01003340 KMQ93645.1 NEVH01018390 PNF23355.1 PYGN01001363 PSN35395.1 GL443495 EFN62032.1 GL887645 EGI70559.1 GBYB01008109 JAG77876.1 ACPB03003193 CH477428 EAT41117.1 GECL01000276 JAP05848.1 GDUN01000309 JAN95610.1 KQ981276 KYN43681.1 KK853306 KDR08705.1 KQ761655 OAD57203.1 KQ982566 KYQ54761.1 KQ977220 KYN04894.1 AY725782 AAU95195.1 GBGD01001849 JAC87040.1 GAPW01002159 JAC11439.1 DS235088 EEB11651.1 KK107419 QOIP01000003 EZA51020.1 RLU25069.1 AF338465 AF338466 AAK25796.1 AAK25797.1 GECZ01006781 JAS62988.1 KQ976407 KYM91399.1 KU296958 ALT55114.1 AY588067 AAT01079.1 GAPW01001631 JAC11967.1 AXCM01000236 KQ971422 EFA12798.2 KF576670 AHH30811.1 ABLF02029597 KQ414596 KOC69925.1 GFXV01003521 MBW15326.1 KC437331 AGK82081.1 ADTU01004257 APCN01002208 KK852680 KDR18593.1 GECZ01029586 JAS40183.1 KC437330 AGK82080.1 ABLF02009131 DQ413235 AK341785 ABD72703.1 BAH72082.1 EEB11648.1 AAAB01008851 EAA07365.5 GGMR01014628 MBY27247.1 KK854500 PTY16384.1 KZ288431 PBC25824.1 JR037914 AEY58032.1 AXCN02000306 AXCN02000307 GGMR01017596 MBY30215.1 ADMH02000313 ETN67016.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000218220
UP000037510
+ More
UP000245037 UP000092462 UP000235965 UP000078492 UP000076502 UP000002358 UP000215335 UP000008237 UP000036403 UP000079169 UP000000311 UP000007755 UP000015103 UP000008820 UP000078541 UP000027135 UP000192223 UP000075809 UP000078542 UP000009046 UP000053097 UP000279307 UP000075902 UP000078540 UP000075883 UP000075881 UP000007266 UP000007819 UP000053825 UP000075882 UP000005205 UP000075840 UP000076407 UP000075901 UP000075903 UP000007062 UP000075885 UP000075920 UP000242457 UP000076408 UP000075886 UP000005203 UP000075900 UP000075880 UP000000673 UP000095300
UP000245037 UP000092462 UP000235965 UP000078492 UP000076502 UP000002358 UP000215335 UP000008237 UP000036403 UP000079169 UP000000311 UP000007755 UP000015103 UP000008820 UP000078541 UP000027135 UP000192223 UP000075809 UP000078542 UP000009046 UP000053097 UP000279307 UP000075902 UP000078540 UP000075883 UP000075881 UP000007266 UP000007819 UP000053825 UP000075882 UP000005205 UP000075840 UP000076407 UP000075901 UP000075903 UP000007062 UP000075885 UP000075920 UP000242457 UP000076408 UP000075886 UP000005203 UP000075900 UP000075880 UP000000673 UP000095300
PRIDE
Pfam
PF00487 FA_desaturase
Interpro
ProteinModelPortal
H9JMC3
A0A212F2M1
A0A194R480
A0A068FRE6
A0A194PH02
B7SB79
+ More
Q8MZZ5 A0A0U2F3D3 A0A0U2F3C0 I7AHL6 A0A2H1VBX7 A0A2A4JFV9 T1RTG7 A0A1L8D6C3 T1RTD2 S4PN29 A0A0M3LAH8 B6CBS4 A0A0L7LIJ1 Q6A4M8 I4DS07 D8UU48 A0A2P8ZGJ8 A0A1B0DIJ1 A0A2J7PNJ8 A0A151JS75 A0A154PDD1 A0A1B6BZA0 K7J0U9 A0A232F3U6 A0A1B6DXZ2 Q4PP96 E2C1U4 A0A0J7NME9 A0A2J7Q458 A0A1S4EHT5 A0A2P8XTS3 E2AX12 F4W5A9 A0A0C9QWJ2 T1HR83 Q173B1 A0A0V0GDS0 A0A0P6J0V5 A0A195FTD4 A0A067QT59 A0A1W4X6D0 A0A310SQM5 A0A151X2W9 A0A195CXM1 Q5XXT0 A0A1S4EFC2 A0A1S3D6B4 A0A069DSW7 A0A023ERW8 E0VE45 A0A1S3D7T3 A0A026W4L1 Q9BH41 A0A1B6GKP5 A0A182TJZ6 A0A195BTK0 A0A1L2BNU6 Q6PPH9 A0A023ERW2 A0A182LZE7 A0A182K1K4 D7EJN1 A0A096XDC8 J9K131 A0A0L7RGE5 A0A182KWY6 A0A2H8TMD9 T1RLC5 A0A158NYT0 A0A182HJS3 A0A182XK65 A0A067RHH4 A0A182T7Z9 A0A182V4X1 A0A1B6ER20 T1RLF5 Q201V1 E0VE42 Q7QDJ5 A0A2S2PCR7 A0A3Q0JL17 A0A182PS55 A0A182W0K0 A0A2R7W9M0 A0A2A3E2X3 V9IBM6 A0A182Y4H9 A0A182QWH3 A0A2S2PLC4 A0A088AI91 A0A182R814 A0A182IPM3 W5JSC3 A0A1I8QDS2
Q8MZZ5 A0A0U2F3D3 A0A0U2F3C0 I7AHL6 A0A2H1VBX7 A0A2A4JFV9 T1RTG7 A0A1L8D6C3 T1RTD2 S4PN29 A0A0M3LAH8 B6CBS4 A0A0L7LIJ1 Q6A4M8 I4DS07 D8UU48 A0A2P8ZGJ8 A0A1B0DIJ1 A0A2J7PNJ8 A0A151JS75 A0A154PDD1 A0A1B6BZA0 K7J0U9 A0A232F3U6 A0A1B6DXZ2 Q4PP96 E2C1U4 A0A0J7NME9 A0A2J7Q458 A0A1S4EHT5 A0A2P8XTS3 E2AX12 F4W5A9 A0A0C9QWJ2 T1HR83 Q173B1 A0A0V0GDS0 A0A0P6J0V5 A0A195FTD4 A0A067QT59 A0A1W4X6D0 A0A310SQM5 A0A151X2W9 A0A195CXM1 Q5XXT0 A0A1S4EFC2 A0A1S3D6B4 A0A069DSW7 A0A023ERW8 E0VE45 A0A1S3D7T3 A0A026W4L1 Q9BH41 A0A1B6GKP5 A0A182TJZ6 A0A195BTK0 A0A1L2BNU6 Q6PPH9 A0A023ERW2 A0A182LZE7 A0A182K1K4 D7EJN1 A0A096XDC8 J9K131 A0A0L7RGE5 A0A182KWY6 A0A2H8TMD9 T1RLC5 A0A158NYT0 A0A182HJS3 A0A182XK65 A0A067RHH4 A0A182T7Z9 A0A182V4X1 A0A1B6ER20 T1RLF5 Q201V1 E0VE42 Q7QDJ5 A0A2S2PCR7 A0A3Q0JL17 A0A182PS55 A0A182W0K0 A0A2R7W9M0 A0A2A3E2X3 V9IBM6 A0A182Y4H9 A0A182QWH3 A0A2S2PLC4 A0A088AI91 A0A182R814 A0A182IPM3 W5JSC3 A0A1I8QDS2
PDB
4YMK
E-value=1.32724e-66,
Score=640
Ontologies
PATHWAY
GO
PANTHER
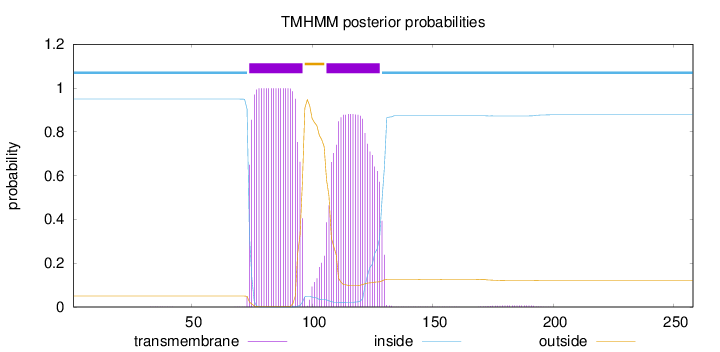
Topology
Length:
258
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
40.41569
Exp number, first 60 AAs:
0.00201
Total prob of N-in:
0.94976
inside
1 - 73
TMhelix
74 - 96
outside
97 - 105
TMhelix
106 - 128
inside
129 - 258
Population Genetic Test Statistics
Pi
4.591881
Theta
7.159303
Tajima's D
0.317802
CLR
11.607398
CSRT
0.486575671216439
Interpretation
Uncertain
