Gene
KWMTBOMO07349
Pre Gene Modal
BGIBMGA010613
Annotation
putative_desaturase_des12?_partial_[Spodoptera_litura]
Location in the cell
PlasmaMembrane Reliability : 3.63
Sequence
CDS
ATGACGACCGGAGCTCATCGACTTTGGGCGCACCGTACTTACAAAGCAAGCACAGAATTGCGCATTGTTCTAATGCTGCTTCAGACTTTAGCTGGTGTTGGCCCTATTTACAACTGGGTTAGACATCACAGACTACATCACGCACACTTTGCTACAGAGAATGATCCATTTGACTACAATCAAGGTTTCGCATACGCCCACATCTTCACGCGAATGAGGAAACTTAGTCCGTATCAGGAAAAATTAAAAGAGTCCATCGACATGTCGGATCTAGAAAATGATTCTGTTGTTATGTTTCAAAAGACGCTATACTGGTTTATGTATGCAACGCTGTTCCTACTGTTGCCCCTGAACGCACCACTCGAGTATTGGGAGGACTCAGTATTAAGTTCCGTCTTTGTTATTGGCTTTCTTCGATACTTCATCTTACTACACGCGTCTTGGTTGATAGAGAGCGCTATCTGCGTTTGGGGATTGAAACCTGGAGAAAAATCACCCCCTGATTCTAATTCTGTGTTCATAATAACGCGAACATTCTGGCCTCACTACCACTATTTAATACCCTATGATTACAAATCCGGAGAATACGGTACCTATGACTCCGGTTGTTCGAGTGCTTTCATTAGAGTTTGGGCAGCACTCGGTATCGCCACAGGTCTTCGTACAGTGGAGACCAATACAATACAGAAGGCTTTGGCTGAGATGGCAAAAACCAAAAAACCTCTTCAAATATGCCTAGAAAATGCCCTAAAAGATCAAAGTTTGTCAGATCAGCATTATCTCAAACGAGAATAA
Protein
MTTGAHRLWAHRTYKASTELRIVLMLLQTLAGVGPIYNWVRHHRLHHAHFATENDPFDYNQGFAYAHIFTRMRKLSPYQEKLKESIDMSDLENDSVVMFQKTLYWFMYATLFLLLPLNAPLEYWEDSVLSSVFVIGFLRYFILLHASWLIESAICVWGLKPGEKSPPDSNSVFIITRTFWPHYHYLIPYDYKSGEYGTYDSGCSSAFIRVWAALGIATGLRTVETNTIQKALAEMAKTKKPLQICLENALKDQSLSDQHYLKRE
Summary
Cofactor
Fe cation
Similarity
Belongs to the fatty acid desaturase type 1 family.
Uniprot
T1RTD5
H9JM61
A0A1V0M870
A0A291P0L9
A0A291P0X2
A0A2A4JJM0
+ More
A0A0P0EA57 A0A0U2F3D5 A0A2H1VUX2 A0A0M3L9C9 A0A068FRE3 A0A212EQ51 A0A194PHZ6 A0A194R3E7 A0A1L8D6P3 A0A1L8D6N2 A0A0L7L1L6 Q17EY5 A0A182LVW9 A0A023EQP9 A0A1S4F571 A0A182GEU2 W5J650 A0A182JYF4 A0A182UMY5 A0A182WRG0 A0A1Q3G5A7 A0A1B6E404 Q7QDC1 A0A084W6J1 A0A336M291 A0A336M071 A0A182W0A1 A0A182HUC0 A0A096XDB8 A0A182IXB6 A0A182FAQ5 A0A182SCW4 D7EIY7 A0A067R818 A0A182N6J2 A0A1I8PP80 A0A182RNM4 T1PHW9 A0A182Y4E6 W8BFR3 W8C5K7 A0A182PAC8 A0A0A1WGL7 A0A0K8UZ01 A0A2P8ZGK2 A0A0K8V658 A0A034WEH4 A0A034WCZ2 A0A182TYR8 C4WWI9 A0A1B6KJ08 A0A1B6I3Q6 A0A1L8DY59 A0A1B6GYT9 T1H7V5 J3JTM2 A0A1B6C828 K7J0V0 B4M6D7 A0A2S2PZ41 A0A1J1I4U0 E0VE43 A0A0G3YLE9 A0A2R7WDH5 B3LZD3 A0A2H8TNW0 A0A1W4W1W3 A0A2S2NXH0 A0A1A9WPD6 D3TNT7 B4GNP5 B5DXR4 A0A0M5J0D3 T1RLH2 A0A023F0M3 B4PLU1 B4JEM8 A0A1A9ZF86 B4HZN6 Q9VA93 T1RLH6 Q4V4B1 B4NGT3 A0A3B0K742 B4R1J9 F5HK89 B3P7W9 A0A0K8V387 B4K9T2 E2C1U0 A0A3B0K759 A0A1B0CEY0 A0A026W4R5
A0A0P0EA57 A0A0U2F3D5 A0A2H1VUX2 A0A0M3L9C9 A0A068FRE3 A0A212EQ51 A0A194PHZ6 A0A194R3E7 A0A1L8D6P3 A0A1L8D6N2 A0A0L7L1L6 Q17EY5 A0A182LVW9 A0A023EQP9 A0A1S4F571 A0A182GEU2 W5J650 A0A182JYF4 A0A182UMY5 A0A182WRG0 A0A1Q3G5A7 A0A1B6E404 Q7QDC1 A0A084W6J1 A0A336M291 A0A336M071 A0A182W0A1 A0A182HUC0 A0A096XDB8 A0A182IXB6 A0A182FAQ5 A0A182SCW4 D7EIY7 A0A067R818 A0A182N6J2 A0A1I8PP80 A0A182RNM4 T1PHW9 A0A182Y4E6 W8BFR3 W8C5K7 A0A182PAC8 A0A0A1WGL7 A0A0K8UZ01 A0A2P8ZGK2 A0A0K8V658 A0A034WEH4 A0A034WCZ2 A0A182TYR8 C4WWI9 A0A1B6KJ08 A0A1B6I3Q6 A0A1L8DY59 A0A1B6GYT9 T1H7V5 J3JTM2 A0A1B6C828 K7J0V0 B4M6D7 A0A2S2PZ41 A0A1J1I4U0 E0VE43 A0A0G3YLE9 A0A2R7WDH5 B3LZD3 A0A2H8TNW0 A0A1W4W1W3 A0A2S2NXH0 A0A1A9WPD6 D3TNT7 B4GNP5 B5DXR4 A0A0M5J0D3 T1RLH2 A0A023F0M3 B4PLU1 B4JEM8 A0A1A9ZF86 B4HZN6 Q9VA93 T1RLH6 Q4V4B1 B4NGT3 A0A3B0K742 B4R1J9 F5HK89 B3P7W9 A0A0K8V387 B4K9T2 E2C1U0 A0A3B0K759 A0A1B0CEY0 A0A026W4R5
Pubmed
24500170
19121390
28349297
28986331
26445454
26385554
+ More
22118469 26354079 26227816 17510324 24945155 26483478 20920257 23761445 12364791 14747013 17210077 24438588 24880119 18362917 19820115 24845553 25315136 25244985 24495485 25830018 29403074 25348373 22516182 23537049 20075255 17994087 20566863 25702953 20353571 15632085 23727612 25474469 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20798317 24508170
22118469 26354079 26227816 17510324 24945155 26483478 20920257 23761445 12364791 14747013 17210077 24438588 24880119 18362917 19820115 24845553 25315136 25244985 24495485 25830018 29403074 25348373 22516182 23537049 20075255 17994087 20566863 25702953 20353571 15632085 23727612 25474469 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20798317 24508170
EMBL
JX531670
AGO45855.1
BABH01033468
KU755475
ARD71185.1
MF687530
+ More
ATJ44456.1 MF687588 ATJ44514.1 NWSH01001345 PCG71600.1 KT261694 ALJ30234.1 KP008126 AKU76413.1 ODYU01004545 SOQ44546.1 KM366788 AIM40218.1 KJ622052 AID66660.1 AGBW02013325 OWR43606.1 KQ459603 KPI92673.1 KQ460845 KPJ12049.1 GEYN01000129 JAV02000.1 GEYN01000125 JAV02004.1 JTDY01003526 KOB69403.1 CH477278 EAT45074.1 AXCM01001395 GAPW01002173 JAC11425.1 JXUM01059125 KQ562036 KXJ76845.1 ADMH02002143 ETN58289.1 GFDL01000057 JAV34988.1 GEDC01024654 GEDC01004657 JAS12644.1 JAS32641.1 AAAB01008859 EAA07943.3 ATLV01020820 KE525308 KFB45835.1 UFQS01000350 UFQT01000350 SSX03095.1 SSX23461.1 SSX03094.1 SSX23460.1 APCN01000618 KF576676 AHH30817.1 KQ971905 EFA12397.2 KK852680 KDR18592.1 KA647468 AFP62097.1 GAMC01008993 GAMC01008989 JAB97562.1 GAMC01008991 JAB97564.1 GBXI01016647 JAC97644.1 GDHF01020551 JAI31763.1 PYGN01000064 PSN55637.1 GDHF01018224 JAI34090.1 GAKP01006402 GAKP01006401 GAKP01006400 JAC52550.1 GAKP01006403 JAC52549.1 ABLF02015065 AK342093 BAH72259.1 GEBQ01028551 JAT11426.1 GECU01026191 JAS81515.1 GFDF01002687 JAV11397.1 GECZ01002197 JAS67572.1 ACPB03020383 BT126579 KB631815 AEE61543.1 ERL86465.1 GEDC01027903 JAS09395.1 CH940652 EDW59213.1 GGMS01001441 MBY70644.1 CVRI01000039 CRK94612.1 DS235088 EEB11649.1 KM507108 AKM28432.1 KK854500 PTY16385.1 CH902617 EDV44112.1 GFXV01003143 MBW14948.1 GGMR01009310 MBY21929.1 CCAG010012086 EZ423089 ADD19365.1 CH479186 EDW39371.1 CM000070 EDY67505.1 CP012526 ALC47548.1 KC437332 AGK82082.1 GBBI01003702 JAC15010.1 CM000160 EDW99078.1 CH916369 EDV93159.1 CH480819 EDW53493.1 AE014297 AAF57021.1 KC437333 AGK82083.1 BT023007 BT023095 BT023167 AAY55423.1 AAY55511.1 AAY55583.1 CH964272 EDW84430.1 OUUW01000041 SPP89935.1 CM000364 EDX14987.1 EGK96641.1 CH954182 EDV53096.1 GDHF01019008 JAI33306.1 CH933806 EDW14557.1 GL452008 EFN78052.1 SPP89934.1 AJWK01009459 KK107419 EZA51018.1
ATJ44456.1 MF687588 ATJ44514.1 NWSH01001345 PCG71600.1 KT261694 ALJ30234.1 KP008126 AKU76413.1 ODYU01004545 SOQ44546.1 KM366788 AIM40218.1 KJ622052 AID66660.1 AGBW02013325 OWR43606.1 KQ459603 KPI92673.1 KQ460845 KPJ12049.1 GEYN01000129 JAV02000.1 GEYN01000125 JAV02004.1 JTDY01003526 KOB69403.1 CH477278 EAT45074.1 AXCM01001395 GAPW01002173 JAC11425.1 JXUM01059125 KQ562036 KXJ76845.1 ADMH02002143 ETN58289.1 GFDL01000057 JAV34988.1 GEDC01024654 GEDC01004657 JAS12644.1 JAS32641.1 AAAB01008859 EAA07943.3 ATLV01020820 KE525308 KFB45835.1 UFQS01000350 UFQT01000350 SSX03095.1 SSX23461.1 SSX03094.1 SSX23460.1 APCN01000618 KF576676 AHH30817.1 KQ971905 EFA12397.2 KK852680 KDR18592.1 KA647468 AFP62097.1 GAMC01008993 GAMC01008989 JAB97562.1 GAMC01008991 JAB97564.1 GBXI01016647 JAC97644.1 GDHF01020551 JAI31763.1 PYGN01000064 PSN55637.1 GDHF01018224 JAI34090.1 GAKP01006402 GAKP01006401 GAKP01006400 JAC52550.1 GAKP01006403 JAC52549.1 ABLF02015065 AK342093 BAH72259.1 GEBQ01028551 JAT11426.1 GECU01026191 JAS81515.1 GFDF01002687 JAV11397.1 GECZ01002197 JAS67572.1 ACPB03020383 BT126579 KB631815 AEE61543.1 ERL86465.1 GEDC01027903 JAS09395.1 CH940652 EDW59213.1 GGMS01001441 MBY70644.1 CVRI01000039 CRK94612.1 DS235088 EEB11649.1 KM507108 AKM28432.1 KK854500 PTY16385.1 CH902617 EDV44112.1 GFXV01003143 MBW14948.1 GGMR01009310 MBY21929.1 CCAG010012086 EZ423089 ADD19365.1 CH479186 EDW39371.1 CM000070 EDY67505.1 CP012526 ALC47548.1 KC437332 AGK82082.1 GBBI01003702 JAC15010.1 CM000160 EDW99078.1 CH916369 EDV93159.1 CH480819 EDW53493.1 AE014297 AAF57021.1 KC437333 AGK82083.1 BT023007 BT023095 BT023167 AAY55423.1 AAY55511.1 AAY55583.1 CH964272 EDW84430.1 OUUW01000041 SPP89935.1 CM000364 EDX14987.1 EGK96641.1 CH954182 EDV53096.1 GDHF01019008 JAI33306.1 CH933806 EDW14557.1 GL452008 EFN78052.1 SPP89934.1 AJWK01009459 KK107419 EZA51018.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000008820 UP000075883 UP000069940 UP000249989 UP000000673 UP000075881 UP000075903 UP000076407 UP000007062 UP000030765 UP000075920 UP000075840 UP000075880 UP000069272 UP000075901 UP000007266 UP000027135 UP000075884 UP000095300 UP000075900 UP000095301 UP000076408 UP000075885 UP000245037 UP000075902 UP000007819 UP000015103 UP000030742 UP000002358 UP000008792 UP000183832 UP000009046 UP000007801 UP000192221 UP000091820 UP000092444 UP000008744 UP000001819 UP000092553 UP000002282 UP000001070 UP000092445 UP000001292 UP000000803 UP000007798 UP000268350 UP000000304 UP000008711 UP000009192 UP000008237 UP000092461 UP000053097
UP000008820 UP000075883 UP000069940 UP000249989 UP000000673 UP000075881 UP000075903 UP000076407 UP000007062 UP000030765 UP000075920 UP000075840 UP000075880 UP000069272 UP000075901 UP000007266 UP000027135 UP000075884 UP000095300 UP000075900 UP000095301 UP000076408 UP000075885 UP000245037 UP000075902 UP000007819 UP000015103 UP000030742 UP000002358 UP000008792 UP000183832 UP000009046 UP000007801 UP000192221 UP000091820 UP000092444 UP000008744 UP000001819 UP000092553 UP000002282 UP000001070 UP000092445 UP000001292 UP000000803 UP000007798 UP000268350 UP000000304 UP000008711 UP000009192 UP000008237 UP000092461 UP000053097
Interpro
Gene 3D
ProteinModelPortal
T1RTD5
H9JM61
A0A1V0M870
A0A291P0L9
A0A291P0X2
A0A2A4JJM0
+ More
A0A0P0EA57 A0A0U2F3D5 A0A2H1VUX2 A0A0M3L9C9 A0A068FRE3 A0A212EQ51 A0A194PHZ6 A0A194R3E7 A0A1L8D6P3 A0A1L8D6N2 A0A0L7L1L6 Q17EY5 A0A182LVW9 A0A023EQP9 A0A1S4F571 A0A182GEU2 W5J650 A0A182JYF4 A0A182UMY5 A0A182WRG0 A0A1Q3G5A7 A0A1B6E404 Q7QDC1 A0A084W6J1 A0A336M291 A0A336M071 A0A182W0A1 A0A182HUC0 A0A096XDB8 A0A182IXB6 A0A182FAQ5 A0A182SCW4 D7EIY7 A0A067R818 A0A182N6J2 A0A1I8PP80 A0A182RNM4 T1PHW9 A0A182Y4E6 W8BFR3 W8C5K7 A0A182PAC8 A0A0A1WGL7 A0A0K8UZ01 A0A2P8ZGK2 A0A0K8V658 A0A034WEH4 A0A034WCZ2 A0A182TYR8 C4WWI9 A0A1B6KJ08 A0A1B6I3Q6 A0A1L8DY59 A0A1B6GYT9 T1H7V5 J3JTM2 A0A1B6C828 K7J0V0 B4M6D7 A0A2S2PZ41 A0A1J1I4U0 E0VE43 A0A0G3YLE9 A0A2R7WDH5 B3LZD3 A0A2H8TNW0 A0A1W4W1W3 A0A2S2NXH0 A0A1A9WPD6 D3TNT7 B4GNP5 B5DXR4 A0A0M5J0D3 T1RLH2 A0A023F0M3 B4PLU1 B4JEM8 A0A1A9ZF86 B4HZN6 Q9VA93 T1RLH6 Q4V4B1 B4NGT3 A0A3B0K742 B4R1J9 F5HK89 B3P7W9 A0A0K8V387 B4K9T2 E2C1U0 A0A3B0K759 A0A1B0CEY0 A0A026W4R5
A0A0P0EA57 A0A0U2F3D5 A0A2H1VUX2 A0A0M3L9C9 A0A068FRE3 A0A212EQ51 A0A194PHZ6 A0A194R3E7 A0A1L8D6P3 A0A1L8D6N2 A0A0L7L1L6 Q17EY5 A0A182LVW9 A0A023EQP9 A0A1S4F571 A0A182GEU2 W5J650 A0A182JYF4 A0A182UMY5 A0A182WRG0 A0A1Q3G5A7 A0A1B6E404 Q7QDC1 A0A084W6J1 A0A336M291 A0A336M071 A0A182W0A1 A0A182HUC0 A0A096XDB8 A0A182IXB6 A0A182FAQ5 A0A182SCW4 D7EIY7 A0A067R818 A0A182N6J2 A0A1I8PP80 A0A182RNM4 T1PHW9 A0A182Y4E6 W8BFR3 W8C5K7 A0A182PAC8 A0A0A1WGL7 A0A0K8UZ01 A0A2P8ZGK2 A0A0K8V658 A0A034WEH4 A0A034WCZ2 A0A182TYR8 C4WWI9 A0A1B6KJ08 A0A1B6I3Q6 A0A1L8DY59 A0A1B6GYT9 T1H7V5 J3JTM2 A0A1B6C828 K7J0V0 B4M6D7 A0A2S2PZ41 A0A1J1I4U0 E0VE43 A0A0G3YLE9 A0A2R7WDH5 B3LZD3 A0A2H8TNW0 A0A1W4W1W3 A0A2S2NXH0 A0A1A9WPD6 D3TNT7 B4GNP5 B5DXR4 A0A0M5J0D3 T1RLH2 A0A023F0M3 B4PLU1 B4JEM8 A0A1A9ZF86 B4HZN6 Q9VA93 T1RLH6 Q4V4B1 B4NGT3 A0A3B0K742 B4R1J9 F5HK89 B3P7W9 A0A0K8V387 B4K9T2 E2C1U0 A0A3B0K759 A0A1B0CEY0 A0A026W4R5
PDB
4YMK
E-value=9.73535e-32,
Score=340
Ontologies
PATHWAY
GO
PANTHER
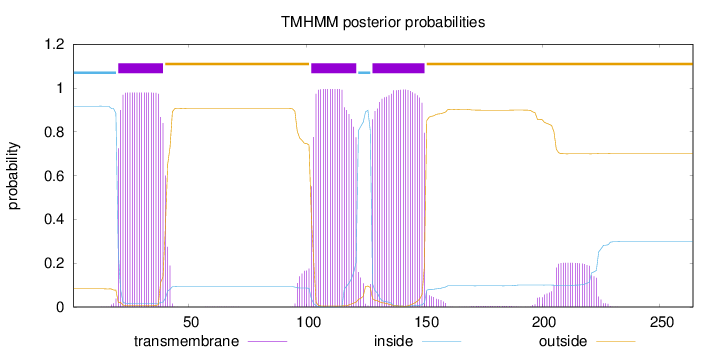
Topology
Length:
264
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
66.36642
Exp number, first 60 AAs:
20.12503
Total prob of N-in:
0.91721
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 39
outside
40 - 101
TMhelix
102 - 121
inside
122 - 127
TMhelix
128 - 150
outside
151 - 264
Population Genetic Test Statistics
Pi
22.465924
Theta
23.125274
Tajima's D
-0.884159
CLR
0.02414
CSRT
0.15949202539873
Interpretation
Uncertain
