Gene
KWMTBOMO07340 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010673
Annotation
PREDICTED:_putative_ATP-dependent_RNA_helicase_me31b_[Bombyx_mori]
Full name
ATP-dependent RNA helicase me31b
Alternative Name
Maternal expression at 31B
Location in the cell
Cytoplasmic Reliability : 1.81 Nuclear Reliability : 2.374
Sequence
CDS
ATGATGACCGAAAATAGAATTAGTTCAAGTAATCACGTCGGAAATAGTATCAGTCAAACTAAAGGAGAGGTCGACAAAAGCATCGACGACGTTGGTTGGAAATCAAAATTAAAAATACCCCCAAAAGACAGAAGAATAAAGACTAGTGATGTCACAGATACTAGAGGCAATGAGTTTGAAGAGTTTTGTTTGAAGCGAGAATTGTTGATGGGTATTTTTGAAAAGGGATGGGAGAAGCCATCTCCTATTCAAGAGGCCTCAATTCCTATTGCCCTAAGTGGAAAAGATGTACTTGCAAGAGCTAAGAATGGAACTGGTAAAACTGGTGCTTATTGTATTCCAGTTTTGGAACAGGTTGATCCTAAAAAGGATACTATACAAGCTTTAATTGTTGTACCAACTAGAGAGTTGGCACTCCAAACATCACAGATTTGTATTGAACTGGCAAAACACACAGACATTCGTGTAATGGTTACTACAGGAGGCACAAACCTTCGAGATGATATAATGCGTATTTATCAAAATGTACAAGTGATTATTGCCACACCTGGGCGTATGATTGATCTCATGGATAAGCAGGTGGCCAAAATGGATCAATGCCGGATGTTAGTTCTTGATGAAGCAGACAAACTTCTTTCACAAGACTTTAAGGGCATGCTGGATATGGTTATTTCAAGATTACCTAAGGAACGCCAAATCTTGCTGTTTTCTGCAACTTTCCCTCTGAGTGTAAAACAATTTATGGAAAAGCATTTGAAAGAACCTTATGAAATAAATCTCATGGAAGAATTAACATTGAAGGGTGTAACACAGTACTATGCATTTGTTCAAGAAAGACAAAAAGTGCATTGCCTTAACACACTTTTTTCAAAGTTGCAAATAAATCAGTCAATAATATTTTGTAATTCAACTCAAAGGGTGGAGTTGTTAGCCAAGAAGATAACAGAGTTAGGATATTGTTGCTACTACATCCACGCACGAATGGCACAAGCTCATCGTAATCGTGTGTTTCACGACTTTCGTGCTGGGCTCTGCCGTAACTTAGTGTGTTCAGACTTGTTCACTAGAGGTATTGATGTGCAAGCAGTTAATGTCGTTATCAATTTTGATTTTCCTCGAATGGCAGAGACATATCTTCATCGCATAGGACGCTCTGGACGTTTTGGGCACCTTGGTATTGCTATAAACTTAATCACATATGATGATCGTTTTGCACTACATCGTATCGAACAAGAACTGGGTACTGAAATCAAGCCAATTCCCAAAGTGATAGATCCTGCGCTATATGTAGCGCGTCCGGAAGATGAAGATCTGGGCGACAAGTAA
Protein
MMTENRISSSNHVGNSISQTKGEVDKSIDDVGWKSKLKIPPKDRRIKTSDVTDTRGNEFEEFCLKRELLMGIFEKGWEKPSPIQEASIPIALSGKDVLARAKNGTGKTGAYCIPVLEQVDPKKDTIQALIVVPTRELALQTSQICIELAKHTDIRVMVTTGGTNLRDDIMRIYQNVQVIIATPGRMIDLMDKQVAKMDQCRMLVLDEADKLLSQDFKGMLDMVISRLPKERQILLFSATFPLSVKQFMEKHLKEPYEINLMEELTLKGVTQYYAFVQERQKVHCLNTLFSKLQINQSIIFCNSTQRVELLAKKITELGYCCYYIHARMAQAHRNRVFHDFRAGLCRNLVCSDLFTRGIDVQAVNVVINFDFPRMAETYLHRIGRSGRFGHLGIAINLITYDDRFALHRIEQELGTEIKPIPKVIDPALYVARPEDEDLGDK
Summary
Description
ATP-dependent RNA helicase which is a core component of a variety of ribonucleoprotein complexes (RNPs) that play critical roles in translational repression and mRNA decapping during embryogenesis, oogenesis, neurogenesis and neurotransmission (PubMed:11546740, PubMed:16256742, PubMed:17178403, PubMed:18590813, PubMed:21267420, PubMed:21447556, PubMed:28875934, PubMed:28388438, PubMed:17982591). Recruits core components and translational repressors to some RNP complexes, and mediates RNP aggregation into processing granules such as P-bodies (PubMed:28875934, PubMed:11546740, PubMed:16256742, PubMed:17178403, PubMed:21267420, PubMed:21447556, PubMed:17982591). As part of a RNP complex containing tral, eIF4E1, cup, and pAbp, involved in RNP-mediated translational repression of maternal mRNAs during oogenesis and embryogenesis (PubMed:28875934). As part of a RNP complex containing tral and the RNA localization factors exu and yps, mediates translational silencing of mRNAs such as osk/oskar and bcd/bicoid during their transport to the oocyte in order to prevent their translation until they reach their positional destinations (PubMed:11546740). In neurons and possibly imaginal disks, involved in miRNA-mediated translational repression, possibly in association with components of the piRNA transposon silencing pathway (PubMed:21447556, PubMed:17178403, PubMed:21267420, PubMed:17982591, PubMed:21081899). Involved in RNA localization and protein trafficking in the oocyte (PubMed:11546740, PubMed:16256742). As part of an ER-associated RNP containing tral, cup and yps, required for tral-dependent ER exit site formation and consequently efficient trafficking of proteins such as grk and yl through the secretory pathway (PubMed:16256742). Component of neuron RNPs that mediate transport and translation of neuronal RNAs, including translation repression of synaptic transcripts in preparation for their dendritic targeting (PubMed:17178403, PubMed:21267420, PubMed:28388438). As part of the Atx2-Not1 repressor complex promotes Not1-dependent post-transcriptional gene silencing in adult circadian pacemaker neurons in order to sustain high-amplitude circadian rhythms and Pdf cycling in a per-independent manner (PubMed:28388438). Promotes the interaction between Atx2 and Not1 within the Atx2-Not1 RNP complex (PubMed:28388438).
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Subunit
Conserved component of different types of multiprotein ribonucleoprotein complexes (RNPs) that form distinct germ granules (P-body, nuage, sponge body or polar granules) and P-body-like neuronal RNPs (PubMed:11546740, PubMed:16256742, PubMed:18765641, PubMed:18590813, PubMed:19285948, PubMed:28388438). Consequently it interacts with a wide variety of proteins, some of which appear to be common interactive partners in almost all RNPs types i.e. cup and tral, whereas other interactions are specific to a germ granule/RNP (PubMed:28945271). Core functional components in me31B-containing RNPs include RNA regulatory proteins (such as translational repressor, RNA-decapping and exonuclease proteins), RNA localization proteins and additional proteins depending on the biological context of the RNPs (PubMed:17178403, PubMed:28945271). In the P-body RNPs, interacts with at least the translation repressor proteins tral, cup and Edc3, and the mRNA localization factor yps (PubMed:16256742, PubMed:18765641, PubMed:18590813, PubMed:19285948). Interaction with tral or Edc3 is required for translation repression and possibly RNA decapping; binding to tral and Edc3 is mutually exclusive (PubMed:18765641, PubMed:19285948). In the nuage and germ plasm polar granule RNPs, interacts with at least tral, cup, and additional proteins required for assembly and function of the germ granules such as tud, vas and aub (PubMed:18765641, PubMed:18590813, PubMed:19285948, PubMed:28945271). Interacts (when dimethylated on Arg residues) with tud; interaction is RNA-independent (PubMed:28945271). Component of the osk RNP complex, which is composed of at least me31B, exu, yps, aret/bruno, cup, and the mRNA of osk (PubMed:10662770). Component of the nos RNP complex, which is composed of at least smg, cup, tral, me31B, the CCR4-NOT complex members Rga/NOT2 and Caf1, and the mRNA of nos (PubMed:21081899). Interacts with tral and piRNA pathway components papi and AGO3; promotes interaction between nuage RNPs and the piRNA-mediated transposon silencing (PubMed:21447556). Forms a RNP containing at least me31B, eIF4E1, cup, tral and pAbp; this interaction is required for the translational silencing of maternal mRNAs during the maternal-to-zygotic transition (PubMed:28875934). In the sponge body, forms a RNP containing at least me31B, exu, yps and the mRNA of osk; interactions with exu and yps are RNA dependent (PubMed:11546740). Component of a neuronal RNP, at least composed of me31B, tral and Fmr1 (PubMed:17178403). Component of the Atx2-Not1 repressor complex, composed of at least me31B, Atx2, tyf and pAbp (PubMed:28388438). Interacts (via the C-terminus) with Atx2, tyf, pAbp and Lsm12 (PubMed:28388438).
Similarity
Belongs to the DEAD box helicase family.
Belongs to the DEAD box helicase family. DDX6/DHH1 subfamily.
Belongs to the DEAD box helicase family. DDX6/DHH1 subfamily.
Keywords
Alternative splicing
ATP-binding
Cell projection
Complete proteome
Cytoplasm
Endoplasmic reticulum
Helicase
Hydrolase
Methylation
Nucleotide-binding
Phosphoprotein
Reference proteome
Repressor
RNA-binding
Translation regulation
Feature
chain ATP-dependent RNA helicase me31b
splice variant In isoform B.
splice variant In isoform B.
Uniprot
A0A2A4IZG5
A0A2H1W8U6
A0A3S2LFJ2
A0A212EQ68
A0A194R3F2
A0A194PHZ1
+ More
S4P9P6 A0A0L7L2C1 A0A2J7PK95 A0A2A3EJ00 V9IH05 A0A087ZPS2 A0A3L8E4W4 A0A154NY43 A0A195ECR5 F4WYD1 A0A195B0L1 A0A195EUR5 A0A158NXN5 A0A151WPT3 A0A151INQ9 K7IWA6 A0A411G6B8 A0A232F259 E2BHT6 E2AXW4 A0A1Y1LM34 A0A026WKK7 A0A0C9R3F6 A0A0T6B766 A0A310SEQ8 A0A0M9A4X6 V5I8P5 A0A0J7KTU2 D2A381 A0A1W4WNE5 A0A2M3Z110 A0A2M4A6Y0 W5JQL4 Q16YL3 A0A182F5D4 A0A182YFB3 A0A1S4H099 A0A182V9Z3 A0A182KAC8 A0A182TSY4 A0A182WYP5 A0A182I526 A0A182NZW5 A0A023ES83 A0A182NF06 A0A182QAX6 A0A1Q3G059 A0A182WH29 A0A084WBY7 A0A182IK09 A0A182R9K9 A0A1L8DS42 A0A182LU81 T1DI54 T1DE88 A0A336KSZ1 A0A2S2QGI1 A0A2H8TP48 Q7PVZ9 A0A336MAM1 X1WJ96 A0A182KUL6 E0VNV1 U5EIR7 A0A0K8VY29 A0A034WY57 A0A0A1WZ36 W8CEF8 H9JMC0 A0A1J1HG36 A0A0L0CF16 B4G7J5 A0A1L8EDM6 N6TLE2 A0A1A9Y6X8 A0A3B0JGW0 A0A1A9UT91 B3MPN5 A0A1A9ZVN5 D3TMT9 A0A1W4UUQ0 B4NZ69 A0A1I8P2L6 B4HWI1 B3N945 B4Q8R4 P23128 T1HZ66 B4LR21 B4KJI4 A0A0P6G5J9 A0A0P4VPZ6 A0A1B0BD30 A0A1A9WLA9
S4P9P6 A0A0L7L2C1 A0A2J7PK95 A0A2A3EJ00 V9IH05 A0A087ZPS2 A0A3L8E4W4 A0A154NY43 A0A195ECR5 F4WYD1 A0A195B0L1 A0A195EUR5 A0A158NXN5 A0A151WPT3 A0A151INQ9 K7IWA6 A0A411G6B8 A0A232F259 E2BHT6 E2AXW4 A0A1Y1LM34 A0A026WKK7 A0A0C9R3F6 A0A0T6B766 A0A310SEQ8 A0A0M9A4X6 V5I8P5 A0A0J7KTU2 D2A381 A0A1W4WNE5 A0A2M3Z110 A0A2M4A6Y0 W5JQL4 Q16YL3 A0A182F5D4 A0A182YFB3 A0A1S4H099 A0A182V9Z3 A0A182KAC8 A0A182TSY4 A0A182WYP5 A0A182I526 A0A182NZW5 A0A023ES83 A0A182NF06 A0A182QAX6 A0A1Q3G059 A0A182WH29 A0A084WBY7 A0A182IK09 A0A182R9K9 A0A1L8DS42 A0A182LU81 T1DI54 T1DE88 A0A336KSZ1 A0A2S2QGI1 A0A2H8TP48 Q7PVZ9 A0A336MAM1 X1WJ96 A0A182KUL6 E0VNV1 U5EIR7 A0A0K8VY29 A0A034WY57 A0A0A1WZ36 W8CEF8 H9JMC0 A0A1J1HG36 A0A0L0CF16 B4G7J5 A0A1L8EDM6 N6TLE2 A0A1A9Y6X8 A0A3B0JGW0 A0A1A9UT91 B3MPN5 A0A1A9ZVN5 D3TMT9 A0A1W4UUQ0 B4NZ69 A0A1I8P2L6 B4HWI1 B3N945 B4Q8R4 P23128 T1HZ66 B4LR21 B4KJI4 A0A0P6G5J9 A0A0P4VPZ6 A0A1B0BD30 A0A1A9WLA9
EC Number
3.6.4.13
Pubmed
22118469
26354079
23622113
26227816
30249741
21719571
+ More
21347285 20075255 28648823 20798317 28004739 24508170 18362917 19820115 20920257 23761445 17510324 25244985 12364791 24945155 26483478 24438588 24330624 20966253 20566863 25348373 25830018 24495485 19121390 26108605 17994087 23537049 20353571 17550304 22936249 1900936 10731132 12537572 12537569 10662770 11546740 16256742 17178403 17982591 18327897 18765641 18590813 19285948 21267420 21447556 21081899 28875934 28945271 28388438 27129103
21347285 20075255 28648823 20798317 28004739 24508170 18362917 19820115 20920257 23761445 17510324 25244985 12364791 24945155 26483478 24438588 24330624 20966253 20566863 25348373 25830018 24495485 19121390 26108605 17994087 23537049 20353571 17550304 22936249 1900936 10731132 12537572 12537569 10662770 11546740 16256742 17178403 17982591 18327897 18765641 18590813 19285948 21267420 21447556 21081899 28875934 28945271 28388438 27129103
EMBL
NWSH01004597
PCG64876.1
ODYU01007045
SOQ49467.1
RSAL01000020
RVE52672.1
+ More
AGBW02013325 OWR43601.1 KQ460845 KPJ12054.1 KQ459603 KPI92668.1 GAIX01003669 JAA88891.1 JTDY01003429 KOB69570.1 NEVH01024940 PNF16761.1 KZ288227 PBC31773.1 JR045312 AEY59952.1 QOIP01000001 RLU27435.1 KQ434782 KZC04589.1 KQ979074 KYN23003.1 GL888440 EGI60783.1 KQ976692 KYM77739.1 KQ981958 KYN31988.1 ADTU01003248 KQ982851 KYQ49876.1 KQ976914 KYN07110.1 MH365626 QBB01435.1 NNAY01001168 OXU24906.1 GL448324 EFN84769.1 GL443736 EFN61728.1 GEZM01051800 GEZM01051799 JAV74693.1 KK107168 EZA56181.1 GBYB01001340 GBYB01001341 JAG71107.1 JAG71108.1 LJIG01009384 KRT83187.1 KQ769958 OAD52719.1 KQ435746 KOX76536.1 GALX01004325 JAB64141.1 LBMM01003169 KMQ93867.1 KQ971338 EFA02274.2 GGFM01001452 MBW22203.1 GGFK01003223 MBW36544.1 ADMH02000797 ETN65039.1 CH477514 EAT39722.1 AAAB01008984 APCN01003750 JXUM01012051 GAPW01001488 KQ560344 JAC12110.1 KXJ82940.1 AXCN02000838 GFDL01001840 JAV33205.1 ATLV01022519 KE525333 KFB47731.1 AXCP01003185 GFDF01004863 JAV09221.1 AXCM01006387 GALA01001136 JAA93716.1 GALA01001137 JAA93715.1 UFQS01000880 UFQT01000880 SSX07467.1 SSX27807.1 GGMS01007644 MBY76847.1 GFXV01003886 MBW15691.1 EAA14695.3 UFQS01000798 UFQT01000798 SSX06981.1 SSX27325.1 ABLF02034022 ABLF02034026 ABLF02034027 DS235354 EEB15057.1 GANO01002524 JAB57347.1 GDHF01008583 JAI43731.1 GAKP01000264 JAC58688.1 GBXI01013005 GBXI01010366 JAD01287.1 JAD03926.1 GAMC01000271 JAC06285.1 BABH01033456 CVRI01000001 CRK86372.1 JRES01000469 KNC30993.1 CH479180 EDW28395.1 GFDG01001981 JAV16818.1 APGK01032042 KB740834 ENN78803.1 OUUW01000006 SPP81634.1 CH902620 EDV32283.1 CCAG010023389 EZ422741 ADD19017.1 CM000157 EDW88764.1 CH480818 EDW52376.1 CH954177 EDV58480.1 CM000361 CM002910 EDX04479.1 KMY89442.1 M59926 AE014134 AY051663 ACPB03008638 CH940649 EDW64560.1 CH933807 EDW13564.1 GDIQ01040699 JAN54038.1 GDKW01001518 JAI55077.1 JXJN01012268
AGBW02013325 OWR43601.1 KQ460845 KPJ12054.1 KQ459603 KPI92668.1 GAIX01003669 JAA88891.1 JTDY01003429 KOB69570.1 NEVH01024940 PNF16761.1 KZ288227 PBC31773.1 JR045312 AEY59952.1 QOIP01000001 RLU27435.1 KQ434782 KZC04589.1 KQ979074 KYN23003.1 GL888440 EGI60783.1 KQ976692 KYM77739.1 KQ981958 KYN31988.1 ADTU01003248 KQ982851 KYQ49876.1 KQ976914 KYN07110.1 MH365626 QBB01435.1 NNAY01001168 OXU24906.1 GL448324 EFN84769.1 GL443736 EFN61728.1 GEZM01051800 GEZM01051799 JAV74693.1 KK107168 EZA56181.1 GBYB01001340 GBYB01001341 JAG71107.1 JAG71108.1 LJIG01009384 KRT83187.1 KQ769958 OAD52719.1 KQ435746 KOX76536.1 GALX01004325 JAB64141.1 LBMM01003169 KMQ93867.1 KQ971338 EFA02274.2 GGFM01001452 MBW22203.1 GGFK01003223 MBW36544.1 ADMH02000797 ETN65039.1 CH477514 EAT39722.1 AAAB01008984 APCN01003750 JXUM01012051 GAPW01001488 KQ560344 JAC12110.1 KXJ82940.1 AXCN02000838 GFDL01001840 JAV33205.1 ATLV01022519 KE525333 KFB47731.1 AXCP01003185 GFDF01004863 JAV09221.1 AXCM01006387 GALA01001136 JAA93716.1 GALA01001137 JAA93715.1 UFQS01000880 UFQT01000880 SSX07467.1 SSX27807.1 GGMS01007644 MBY76847.1 GFXV01003886 MBW15691.1 EAA14695.3 UFQS01000798 UFQT01000798 SSX06981.1 SSX27325.1 ABLF02034022 ABLF02034026 ABLF02034027 DS235354 EEB15057.1 GANO01002524 JAB57347.1 GDHF01008583 JAI43731.1 GAKP01000264 JAC58688.1 GBXI01013005 GBXI01010366 JAD01287.1 JAD03926.1 GAMC01000271 JAC06285.1 BABH01033456 CVRI01000001 CRK86372.1 JRES01000469 KNC30993.1 CH479180 EDW28395.1 GFDG01001981 JAV16818.1 APGK01032042 KB740834 ENN78803.1 OUUW01000006 SPP81634.1 CH902620 EDV32283.1 CCAG010023389 EZ422741 ADD19017.1 CM000157 EDW88764.1 CH480818 EDW52376.1 CH954177 EDV58480.1 CM000361 CM002910 EDX04479.1 KMY89442.1 M59926 AE014134 AY051663 ACPB03008638 CH940649 EDW64560.1 CH933807 EDW13564.1 GDIQ01040699 JAN54038.1 GDKW01001518 JAI55077.1 JXJN01012268
Proteomes
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000235965 UP000242457 UP000005203 UP000279307 UP000076502 UP000078492 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000078542 UP000002358 UP000215335 UP000008237 UP000000311 UP000053097 UP000053105 UP000036403 UP000007266 UP000192223 UP000000673 UP000008820 UP000069272 UP000076408 UP000075903 UP000075881 UP000075902 UP000076407 UP000075840 UP000075885 UP000069940 UP000249989 UP000075884 UP000075886 UP000075920 UP000030765 UP000075880 UP000075900 UP000075883 UP000007062 UP000007819 UP000075882 UP000009046 UP000005204 UP000183832 UP000037069 UP000008744 UP000019118 UP000092443 UP000268350 UP000078200 UP000007801 UP000092445 UP000092444 UP000192221 UP000002282 UP000095300 UP000001292 UP000008711 UP000000304 UP000000803 UP000015103 UP000008792 UP000009192 UP000092460 UP000091820
UP000235965 UP000242457 UP000005203 UP000279307 UP000076502 UP000078492 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000078542 UP000002358 UP000215335 UP000008237 UP000000311 UP000053097 UP000053105 UP000036403 UP000007266 UP000192223 UP000000673 UP000008820 UP000069272 UP000076408 UP000075903 UP000075881 UP000075902 UP000076407 UP000075840 UP000075885 UP000069940 UP000249989 UP000075884 UP000075886 UP000075920 UP000030765 UP000075880 UP000075900 UP000075883 UP000007062 UP000007819 UP000075882 UP000009046 UP000005204 UP000183832 UP000037069 UP000008744 UP000019118 UP000092443 UP000268350 UP000078200 UP000007801 UP000092445 UP000092444 UP000192221 UP000002282 UP000095300 UP000001292 UP000008711 UP000000304 UP000000803 UP000015103 UP000008792 UP000009192 UP000092460 UP000091820
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
A0A2A4IZG5
A0A2H1W8U6
A0A3S2LFJ2
A0A212EQ68
A0A194R3F2
A0A194PHZ1
+ More
S4P9P6 A0A0L7L2C1 A0A2J7PK95 A0A2A3EJ00 V9IH05 A0A087ZPS2 A0A3L8E4W4 A0A154NY43 A0A195ECR5 F4WYD1 A0A195B0L1 A0A195EUR5 A0A158NXN5 A0A151WPT3 A0A151INQ9 K7IWA6 A0A411G6B8 A0A232F259 E2BHT6 E2AXW4 A0A1Y1LM34 A0A026WKK7 A0A0C9R3F6 A0A0T6B766 A0A310SEQ8 A0A0M9A4X6 V5I8P5 A0A0J7KTU2 D2A381 A0A1W4WNE5 A0A2M3Z110 A0A2M4A6Y0 W5JQL4 Q16YL3 A0A182F5D4 A0A182YFB3 A0A1S4H099 A0A182V9Z3 A0A182KAC8 A0A182TSY4 A0A182WYP5 A0A182I526 A0A182NZW5 A0A023ES83 A0A182NF06 A0A182QAX6 A0A1Q3G059 A0A182WH29 A0A084WBY7 A0A182IK09 A0A182R9K9 A0A1L8DS42 A0A182LU81 T1DI54 T1DE88 A0A336KSZ1 A0A2S2QGI1 A0A2H8TP48 Q7PVZ9 A0A336MAM1 X1WJ96 A0A182KUL6 E0VNV1 U5EIR7 A0A0K8VY29 A0A034WY57 A0A0A1WZ36 W8CEF8 H9JMC0 A0A1J1HG36 A0A0L0CF16 B4G7J5 A0A1L8EDM6 N6TLE2 A0A1A9Y6X8 A0A3B0JGW0 A0A1A9UT91 B3MPN5 A0A1A9ZVN5 D3TMT9 A0A1W4UUQ0 B4NZ69 A0A1I8P2L6 B4HWI1 B3N945 B4Q8R4 P23128 T1HZ66 B4LR21 B4KJI4 A0A0P6G5J9 A0A0P4VPZ6 A0A1B0BD30 A0A1A9WLA9
S4P9P6 A0A0L7L2C1 A0A2J7PK95 A0A2A3EJ00 V9IH05 A0A087ZPS2 A0A3L8E4W4 A0A154NY43 A0A195ECR5 F4WYD1 A0A195B0L1 A0A195EUR5 A0A158NXN5 A0A151WPT3 A0A151INQ9 K7IWA6 A0A411G6B8 A0A232F259 E2BHT6 E2AXW4 A0A1Y1LM34 A0A026WKK7 A0A0C9R3F6 A0A0T6B766 A0A310SEQ8 A0A0M9A4X6 V5I8P5 A0A0J7KTU2 D2A381 A0A1W4WNE5 A0A2M3Z110 A0A2M4A6Y0 W5JQL4 Q16YL3 A0A182F5D4 A0A182YFB3 A0A1S4H099 A0A182V9Z3 A0A182KAC8 A0A182TSY4 A0A182WYP5 A0A182I526 A0A182NZW5 A0A023ES83 A0A182NF06 A0A182QAX6 A0A1Q3G059 A0A182WH29 A0A084WBY7 A0A182IK09 A0A182R9K9 A0A1L8DS42 A0A182LU81 T1DI54 T1DE88 A0A336KSZ1 A0A2S2QGI1 A0A2H8TP48 Q7PVZ9 A0A336MAM1 X1WJ96 A0A182KUL6 E0VNV1 U5EIR7 A0A0K8VY29 A0A034WY57 A0A0A1WZ36 W8CEF8 H9JMC0 A0A1J1HG36 A0A0L0CF16 B4G7J5 A0A1L8EDM6 N6TLE2 A0A1A9Y6X8 A0A3B0JGW0 A0A1A9UT91 B3MPN5 A0A1A9ZVN5 D3TMT9 A0A1W4UUQ0 B4NZ69 A0A1I8P2L6 B4HWI1 B3N945 B4Q8R4 P23128 T1HZ66 B4LR21 B4KJI4 A0A0P6G5J9 A0A0P4VPZ6 A0A1B0BD30 A0A1A9WLA9
PDB
5ANR
E-value=7.52071e-176,
Score=1585
Ontologies
GO
GO:0003676
GO:0004386
GO:0005524
GO:0003743
GO:0035195
GO:2000766
GO:0050688
GO:1990124
GO:0000932
GO:1905618
GO:0043186
GO:0070971
GO:0043025
GO:0098794
GO:1990120
GO:0071683
GO:0090328
GO:0071598
GO:0003723
GO:0030707
GO:0033962
GO:0150011
GO:0007279
GO:0046959
GO:0060148
GO:0005737
GO:0036464
GO:0005783
GO:0043565
GO:0005852
GO:0006446
GO:0043022
GO:0005488
GO:0003707
GO:0051537
Topology
Subcellular location
Cytoplasm
Component of a variety of germ granule ribonucleoprotein complexes (RNPs) including the nuage of nurse cells, sponge bodies of nurse cells and early egg chambers in oocytes, as well as polar granules in the germ plasm at the posterior pole of mid-late stage oocytes and in early embryos (PubMed:11546740, PubMed:18590813, PubMed:21447556, PubMed:28945271, PubMed:21267420). Component of an RNP that localizes to discrete subdomains of the ER cytoplasmic surface (PubMed:21267420, PubMed:16256742). Also present in cytoplasmic granules in the cell bodies and neuropil area of the antennal lobes (PubMed:21267420). In the olfactory sensory and projection neurons, granules appear to predominately localize to postsynaptic dendrites (PubMed:21267420). With evidence from 5 publications.
Cytoplasmic ribonucleoprotein granule Component of a variety of germ granule ribonucleoprotein complexes (RNPs) including the nuage of nurse cells, sponge bodies of nurse cells and early egg chambers in oocytes, as well as polar granules in the germ plasm at the posterior pole of mid-late stage oocytes and in early embryos (PubMed:11546740, PubMed:18590813, PubMed:21447556, PubMed:28945271, PubMed:21267420). Component of an RNP that localizes to discrete subdomains of the ER cytoplasmic surface (PubMed:21267420, PubMed:16256742). Also present in cytoplasmic granules in the cell bodies and neuropil area of the antennal lobes (PubMed:21267420). In the olfactory sensory and projection neurons, granules appear to predominately localize to postsynaptic dendrites (PubMed:21267420). With evidence from 5 publications.
P-body Component of a variety of germ granule ribonucleoprotein complexes (RNPs) including the nuage of nurse cells, sponge bodies of nurse cells and early egg chambers in oocytes, as well as polar granules in the germ plasm at the posterior pole of mid-late stage oocytes and in early embryos (PubMed:11546740, PubMed:18590813, PubMed:21447556, PubMed:28945271, PubMed:21267420). Component of an RNP that localizes to discrete subdomains of the ER cytoplasmic surface (PubMed:21267420, PubMed:16256742). Also present in cytoplasmic granules in the cell bodies and neuropil area of the antennal lobes (PubMed:21267420). In the olfactory sensory and projection neurons, granules appear to predominately localize to postsynaptic dendrites (PubMed:21267420). With evidence from 5 publications.
Endoplasmic reticulum Component of a variety of germ granule ribonucleoprotein complexes (RNPs) including the nuage of nurse cells, sponge bodies of nurse cells and early egg chambers in oocytes, as well as polar granules in the germ plasm at the posterior pole of mid-late stage oocytes and in early embryos (PubMed:11546740, PubMed:18590813, PubMed:21447556, PubMed:28945271, PubMed:21267420). Component of an RNP that localizes to discrete subdomains of the ER cytoplasmic surface (PubMed:21267420, PubMed:16256742). Also present in cytoplasmic granules in the cell bodies and neuropil area of the antennal lobes (PubMed:21267420). In the olfactory sensory and projection neurons, granules appear to predominately localize to postsynaptic dendrites (PubMed:21267420). With evidence from 5 publications.
Cell projection Component of a variety of germ granule ribonucleoprotein complexes (RNPs) including the nuage of nurse cells, sponge bodies of nurse cells and early egg chambers in oocytes, as well as polar granules in the germ plasm at the posterior pole of mid-late stage oocytes and in early embryos (PubMed:11546740, PubMed:18590813, PubMed:21447556, PubMed:28945271, PubMed:21267420). Component of an RNP that localizes to discrete subdomains of the ER cytoplasmic surface (PubMed:21267420, PubMed:16256742). Also present in cytoplasmic granules in the cell bodies and neuropil area of the antennal lobes (PubMed:21267420). In the olfactory sensory and projection neurons, granules appear to predominately localize to postsynaptic dendrites (PubMed:21267420). With evidence from 5 publications.
Dendrite Component of a variety of germ granule ribonucleoprotein complexes (RNPs) including the nuage of nurse cells, sponge bodies of nurse cells and early egg chambers in oocytes, as well as polar granules in the germ plasm at the posterior pole of mid-late stage oocytes and in early embryos (PubMed:11546740, PubMed:18590813, PubMed:21447556, PubMed:28945271, PubMed:21267420). Component of an RNP that localizes to discrete subdomains of the ER cytoplasmic surface (PubMed:21267420, PubMed:16256742). Also present in cytoplasmic granules in the cell bodies and neuropil area of the antennal lobes (PubMed:21267420). In the olfactory sensory and projection neurons, granules appear to predominately localize to postsynaptic dendrites (PubMed:21267420). With evidence from 5 publications.
Cytoplasmic ribonucleoprotein granule Component of a variety of germ granule ribonucleoprotein complexes (RNPs) including the nuage of nurse cells, sponge bodies of nurse cells and early egg chambers in oocytes, as well as polar granules in the germ plasm at the posterior pole of mid-late stage oocytes and in early embryos (PubMed:11546740, PubMed:18590813, PubMed:21447556, PubMed:28945271, PubMed:21267420). Component of an RNP that localizes to discrete subdomains of the ER cytoplasmic surface (PubMed:21267420, PubMed:16256742). Also present in cytoplasmic granules in the cell bodies and neuropil area of the antennal lobes (PubMed:21267420). In the olfactory sensory and projection neurons, granules appear to predominately localize to postsynaptic dendrites (PubMed:21267420). With evidence from 5 publications.
P-body Component of a variety of germ granule ribonucleoprotein complexes (RNPs) including the nuage of nurse cells, sponge bodies of nurse cells and early egg chambers in oocytes, as well as polar granules in the germ plasm at the posterior pole of mid-late stage oocytes and in early embryos (PubMed:11546740, PubMed:18590813, PubMed:21447556, PubMed:28945271, PubMed:21267420). Component of an RNP that localizes to discrete subdomains of the ER cytoplasmic surface (PubMed:21267420, PubMed:16256742). Also present in cytoplasmic granules in the cell bodies and neuropil area of the antennal lobes (PubMed:21267420). In the olfactory sensory and projection neurons, granules appear to predominately localize to postsynaptic dendrites (PubMed:21267420). With evidence from 5 publications.
Endoplasmic reticulum Component of a variety of germ granule ribonucleoprotein complexes (RNPs) including the nuage of nurse cells, sponge bodies of nurse cells and early egg chambers in oocytes, as well as polar granules in the germ plasm at the posterior pole of mid-late stage oocytes and in early embryos (PubMed:11546740, PubMed:18590813, PubMed:21447556, PubMed:28945271, PubMed:21267420). Component of an RNP that localizes to discrete subdomains of the ER cytoplasmic surface (PubMed:21267420, PubMed:16256742). Also present in cytoplasmic granules in the cell bodies and neuropil area of the antennal lobes (PubMed:21267420). In the olfactory sensory and projection neurons, granules appear to predominately localize to postsynaptic dendrites (PubMed:21267420). With evidence from 5 publications.
Cell projection Component of a variety of germ granule ribonucleoprotein complexes (RNPs) including the nuage of nurse cells, sponge bodies of nurse cells and early egg chambers in oocytes, as well as polar granules in the germ plasm at the posterior pole of mid-late stage oocytes and in early embryos (PubMed:11546740, PubMed:18590813, PubMed:21447556, PubMed:28945271, PubMed:21267420). Component of an RNP that localizes to discrete subdomains of the ER cytoplasmic surface (PubMed:21267420, PubMed:16256742). Also present in cytoplasmic granules in the cell bodies and neuropil area of the antennal lobes (PubMed:21267420). In the olfactory sensory and projection neurons, granules appear to predominately localize to postsynaptic dendrites (PubMed:21267420). With evidence from 5 publications.
Dendrite Component of a variety of germ granule ribonucleoprotein complexes (RNPs) including the nuage of nurse cells, sponge bodies of nurse cells and early egg chambers in oocytes, as well as polar granules in the germ plasm at the posterior pole of mid-late stage oocytes and in early embryos (PubMed:11546740, PubMed:18590813, PubMed:21447556, PubMed:28945271, PubMed:21267420). Component of an RNP that localizes to discrete subdomains of the ER cytoplasmic surface (PubMed:21267420, PubMed:16256742). Also present in cytoplasmic granules in the cell bodies and neuropil area of the antennal lobes (PubMed:21267420). In the olfactory sensory and projection neurons, granules appear to predominately localize to postsynaptic dendrites (PubMed:21267420). With evidence from 5 publications.
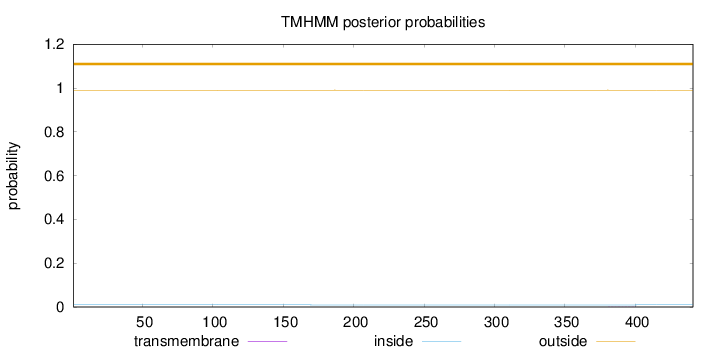
Length:
441
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01117
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00975
outside
1 - 441
Population Genetic Test Statistics
Pi
2.177638
Theta
5.778261
Tajima's D
-1.468216
CLR
0.887207
CSRT
0.0582470876456177
Interpretation
Uncertain
