Gene
KWMTBOMO07333
Annotation
PREDICTED:_WASH_complex_subunit_7-like_[Plutella_xylostella]
Full name
WASH complex subunit 4
Alternative Name
Strumpellin and WASH-interacting protein
WASH complex subunit SWIP
WASH complex subunit SWIP
Location in the cell
Nuclear Reliability : 2.012 PlasmaMembrane Reliability : 1.831
Sequence
CDS
ATGAAAAGTGATTTCGAGAAAGAAAGTACAAGCAATATTTTAAAAAAATATGGCATATTTTTCAAGATGCAGTATCAGGTTTTGGCATTCAATTGCAATAATGAAATACTAGATGATAATAAAACATTTTTGCCGATTCAATTACTCATGAGTCCACACGAGCTCGTACCAATTACTGAGTTAGTAGTATCGGATAACACATTTATGACTAAAGTTTTGAGTGTGTTTACTTCCTTATGTGTTGAAGTAAATAACTTGAAAGAGGAAGCCTTTGAAAAGTATTTTATCTACTTGGCTGCTTATGAAAAATATGAAACAGAAGATGTGAGTTCCCAGATGGAAATGATATGCTTACTCTGCGCATTTATCATTCGATGTGATGAAACTTTAAGAAACATGTGCAACCAAGTTTTTGCTTTGTTTTCCGATGGAACTGTTAATCTCAATTGCATTCAACTACACTATATCATCAACCACATTGGAGAGATATTTACTATTCTTGTCTTACTTGAAATACTGATAGCCAATACTTCAATGCCCACAAATTGGAATAAATTTTGCAAAACATTAAAAACATACCATAACAAGAACTCGGCAGCTTTAGATATTACTGAAGAGAAATTTAATGCAATGATGGGGGCTATTAATAACATCACAGTCAAAATAATGAATGATGATATTGTACAAAAATCATTACAAAGTGTATCACTATTGAGAAAGAACTTGATTGACAAGAATTGTTCTATGATAGCAACAGAGTTTTCACACTACCTTAAACAGGGAATTATAAATTTGGACAAACTTGTACAGGAACAACCCAATGTTCGAAATATGTACAAATGTATTAAAATTAATAGTTTATTTGTATTGTCTTCACAATTGTTTGGTAATGTAGACAAGAAGATATTTAAGTCATTAGTGGAATTGAACACAAAGGCACACAGTGTTCATGTGATTGGAACAACTATGTGGTATCCTGAGCAATTTCTACAGAAACATGCCCCTGCACTTTGTGCAAAACAGGAAAAATTATCACAAACTATGTTGAAAACACGGCAATCGTATATCAATATGAAAAAAACTAGCCTATCTCGTGATATAATTAATCTTCAAATAGTATGTGGTCAATGGGTGTTAAATATTGAAGATTTCTTTTCTGCTGATCATCATAGGCTTAGTGCTGCTGAAATGAATTTACATTCCAGAACTCTTTTAAATGGCTTGGACCTAGCATCCAGAATCAATCATGAAGTTTTGACATTGATAAATTTACATCTTTCCCTTGGGATACCTCTTCCAAAGAGTATGTTAATGTTGGTATTTGAGATAATCAATATTTTAAAATCAATGCTGAATGCAATATCACGGCATAGAAGCCAAATAATTTTATCTATCAATATGGTCATTCAACATTTTGTATTCCAAGCAACATCAACAATATTAGAAGTAAAGAAATTATTAGTTTCAGACAAGAATTTCGCAAATAAAAGATTGGATGAACTTACATGCATCGTTATTGCAGAAAAAGCACTTCGTGGCGTAGCTACAATTGAACGAAATGTAGTAGCTAACACAGCACTGAGCTTTATACCTGAAAGTACTTATTTAGAAGACAGCTATGTAAAGCTTGGTTTGTTGTTAGAGAATGTTCAAACATTGACAAACTTGATGAAATCAATGGAAAGTTTTTGCAATTGCTCTTGGCTGTTCTGGCATCAGAACATATTTCCAATTTACTTTGATCAAAACTTTTCTATGCAATTGAGTACTTTAAAATTAAAATACTTCTTAATGGCCTTAGAAGACTGTACGGTGCTATTACATAAAACAGATAAATTATTTGATAGTAATATAAAAGAAGATTTTATGGAAACAGTTAAAAAGTACATGGAACAAAAATTTATCAAAGGTGCAAGTCAGAATATTGAAACAAATTTACGATTGCATATACATTCACATCTTCAACTTGATGTAATCAATCCATTTACTTTTGAAATGGTAAAAAAATTGCTTCTAGCAATTAATACTTTTAGAGTTCAGTACATTTACATGTGTGTTGTGCCATCAGTTGAACACTATCTTTCAACAATGTTCTACAACCTAACTACCGTTGTATTATCAGATTGGAAAACTTATGGTGAAATGAGACAAATGGCGAGACTAAAATTCAATTTAAAAACGGTTCAAGATAATCTCCCAACTCAGACTCTGGAGCAGGGCCTAGATATACTAGAGATTATGCGCAACATTCATATATTCGTTTCGAAGTATCTTTACAATCTAAACAATCAAATATTTATTGAAAGAAGCAGCAACAATAAACACTTGAGCTCAATTAACATTCGACATGTCGCCAATTCTATACGGACTCATGGTACTGGGATAATGAACACAACAGTGAATTTTACATACCAGTTTTTAAAAAAGAAGTTCTTTACATTTTCTCAGTTCATGTATGATGAACATATAAAATCCAGGTTAATTAAAGATCTTAGAATCTTCAAAGAAACCACTGATGGAAACTACAGCTACATGTATCCATTTAAAAATGCAGAAAGATTTAACAAGGGTATAAAGGTGTTGGGACTTTCAGAGGATGGACAGAGTTACTTAGATCTGTTTCGAGATCTTATTAGTCAAATTGGAAACGCCATGGGATATGTTAGGATGATTAGGTCAGGCGGCCGACATTGCTGTTCGGATGCGACTGTTTTCCTGCCAAATCTGGAAGATGTTGTTAATTTCAAAGAACTATGCCAAAAAGAGGCTCTTGGGCCCCAGACTGTTCAATCAGCGGAAATTCTAGATCAGAACACAAACGACCTTATTTCCAACTTTATTCAAGGAACTGAATATTTCAAGCTGCTGGTAGACGTGTTCTCACCCGTTTTCCGAAATCCTAAGAATGTTCATTTGAAGAATTTTTATATTATTGTTCCTCCATTGACACTTAATTTTGTGGAACATCTAATCTTGTCGAAAGATAAAATGTCGAAAAAGAATAAAATAGGAGCTGCTTTTACTGATGATGGTTTTGCTATGGGAATTGCTTACATACTGAAATTACTAGATCAGGATTCTAATTTTGAATCTCTACACTGGTTTGAATCAGTATGGAAACACATTGATGACGAAAGAAAGTTACTAGAAGAACAGAAAAACAAAGGAACCATTCAATTACAACAAGCCTTAGCATTGACCGAGAAGAAAATAAAAACATTAGAAGAAGAGTACAAACTCCTCTATTACAGTCTTACAAGTGCCAGAATATTTTTTCGTTAA
Protein
MKSDFEKESTSNILKKYGIFFKMQYQVLAFNCNNEILDDNKTFLPIQLLMSPHELVPITELVVSDNTFMTKVLSVFTSLCVEVNNLKEEAFEKYFIYLAAYEKYETEDVSSQMEMICLLCAFIIRCDETLRNMCNQVFALFSDGTVNLNCIQLHYIINHIGEIFTILVLLEILIANTSMPTNWNKFCKTLKTYHNKNSAALDITEEKFNAMMGAINNITVKIMNDDIVQKSLQSVSLLRKNLIDKNCSMIATEFSHYLKQGIINLDKLVQEQPNVRNMYKCIKINSLFVLSSQLFGNVDKKIFKSLVELNTKAHSVHVIGTTMWYPEQFLQKHAPALCAKQEKLSQTMLKTRQSYINMKKTSLSRDIINLQIVCGQWVLNIEDFFSADHHRLSAAEMNLHSRTLLNGLDLASRINHEVLTLINLHLSLGIPLPKSMLMLVFEIINILKSMLNAISRHRSQIILSINMVIQHFVFQATSTILEVKKLLVSDKNFANKRLDELTCIVIAEKALRGVATIERNVVANTALSFIPESTYLEDSYVKLGLLLENVQTLTNLMKSMESFCNCSWLFWHQNIFPIYFDQNFSMQLSTLKLKYFLMALEDCTVLLHKTDKLFDSNIKEDFMETVKKYMEQKFIKGASQNIETNLRLHIHSHLQLDVINPFTFEMVKKLLLAINTFRVQYIYMCVVPSVEHYLSTMFYNLTTVVLSDWKTYGEMRQMARLKFNLKTVQDNLPTQTLEQGLDILEIMRNIHIFVSKYLYNLNNQIFIERSSNNKHLSSINIRHVANSIRTHGTGIMNTTVNFTYQFLKKKFFTFSQFMYDEHIKSRLIKDLRIFKETTDGNYSYMYPFKNAERFNKGIKVLGLSEDGQSYLDLFRDLISQIGNAMGYVRMIRSGGRHCCSDATVFLPNLEDVVNFKELCQKEALGPQTVQSAEILDQNTNDLISNFIQGTEYFKLLVDVFSPVFRNPKNVHLKNFYIIVPPLTLNFVEHLILSKDKMSKKNKIGAAFTDDGFAMGIAYILKLLDQDSNFESLHWFESVWKHIDDERKLLEEQKNKGTIQLQQALALTEKKIKTLEEEYKLLYYSLTSARIFFR
Summary
Description
Acts at least in part as component of the WASH core complex whose assembly at the surface of endosomes seems to inhibit WASH nucleation-promoting factor (NPF) activity in recruiting and activating the Arp2/3 complex to induce actin polymerization, and which is involved in the regulation of the fission of tubules that serve as transport intermediates during endosome sorting (PubMed:19922875, PubMed:20498093).
Subunit
Component of the WASH core complex also described as WASH regulatory complex (SHRC) composed of WASH (WASHC1, WASH2P or WASH3P), WASHC2 (WASHC2A or WASHC2C), WASHC3, WASHC4 and WASHC5. The WASH core complex associates via WASHC2 with the F-actin-capping protein dimer (formed by CAPZA1, CAPZA2 or CAPZA3 and CAPZB) in a transient or substoichiometric manner which was initially described as WASH complex.
Similarity
Belongs to the SWIP family.
Keywords
Acetylation
Alternative splicing
Coiled coil
Complete proteome
Disease mutation
Endosome
Mental retardation
Phosphoprotein
Polymorphism
Protein transport
Reference proteome
Transport
Feature
chain WASH complex subunit 4
splice variant In isoform 2.
sequence variant In dbSNP:rs34434425.
splice variant In isoform 2.
sequence variant In dbSNP:rs34434425.
Uniprot
A0A2H1VDY1
A0A2A4JWE3
A0A1E1W053
A0A3S2LFI3
A0A194PIW6
A0A194R2Z4
+ More
A0A212ETZ5 A0A0L7KRI2 A0A1E1WK11 A0A067QYP0 A0A2J7PXZ5 A0A210QJ49 A0A2J7PXZ8 T1J3X6 A0A1S3JFP0 H0ZLG1 L5KF43 M3VK14 A0A3Q2IAN7 G5E600 L8IEP9 I3LLV0 H0WQT5 A0A2C9JNT2 R4GHH2 A0A1V4K7W3 A0A093RRJ7 W5Q6S3 M3VWP4 G1P956 A0A341AB65 A0A2U3ZZ47 A0A2Y9QKE1 A0A224YRZ8 A0A091Q5C7 A0A091P062 U3EJC9 A0A131Z8M7 R7UQC0 U3KG72 F7HA23 A0A3Q7WYU4 A0A091GNN6 D2H1T9 A0A1S3ACV0 A0A2U3WXU5 A0A383YSH6 A0A091UID3 A0A2K5F184 A0A2K6UPC6 G1NJT1 A0A3Q7PT10 A0A2K5RWX0 A0A2Y9H0S5 A0A3Q7Q7T4 A0A091EW44 A0A094LF41 J9P227 A0A2Y9T2K3 A0A2Y9IU07 A0A2C9JNS4 A0A2C9JNS6 A0A091G738 G3W5X6 A0A3Q7TN06 H2R755 A0A131Y0E5 C3YUG4 G5C464 A0A2R9CAM4 A0A2J8XM21 A0A2Y9IRC4 M3YL02 A0A3L8SCM6 H0V7S3 G7N5B6 H9EPA4 L7M974 A0A094K1P4 A0A384DAB7 A0A2U3ZUV1 Q2M389 A0A093D5J2 A0A1W7RGE6 A0A091QTV7 A0A2K5N2C0 A0A2Y9DFV9 B7PRX4 A0A091S5I5 I3MMP7 A0A2K6BLP5 G7PJE1 A0A2K5VNY9 A0A0D9S2H4 A0A096MN14 A0A2K5N1T9 A0A091I6Q2 A0A2Y9IY17 A0A2J8MZR6 A0A131XDK8
A0A212ETZ5 A0A0L7KRI2 A0A1E1WK11 A0A067QYP0 A0A2J7PXZ5 A0A210QJ49 A0A2J7PXZ8 T1J3X6 A0A1S3JFP0 H0ZLG1 L5KF43 M3VK14 A0A3Q2IAN7 G5E600 L8IEP9 I3LLV0 H0WQT5 A0A2C9JNT2 R4GHH2 A0A1V4K7W3 A0A093RRJ7 W5Q6S3 M3VWP4 G1P956 A0A341AB65 A0A2U3ZZ47 A0A2Y9QKE1 A0A224YRZ8 A0A091Q5C7 A0A091P062 U3EJC9 A0A131Z8M7 R7UQC0 U3KG72 F7HA23 A0A3Q7WYU4 A0A091GNN6 D2H1T9 A0A1S3ACV0 A0A2U3WXU5 A0A383YSH6 A0A091UID3 A0A2K5F184 A0A2K6UPC6 G1NJT1 A0A3Q7PT10 A0A2K5RWX0 A0A2Y9H0S5 A0A3Q7Q7T4 A0A091EW44 A0A094LF41 J9P227 A0A2Y9T2K3 A0A2Y9IU07 A0A2C9JNS4 A0A2C9JNS6 A0A091G738 G3W5X6 A0A3Q7TN06 H2R755 A0A131Y0E5 C3YUG4 G5C464 A0A2R9CAM4 A0A2J8XM21 A0A2Y9IRC4 M3YL02 A0A3L8SCM6 H0V7S3 G7N5B6 H9EPA4 L7M974 A0A094K1P4 A0A384DAB7 A0A2U3ZUV1 Q2M389 A0A093D5J2 A0A1W7RGE6 A0A091QTV7 A0A2K5N2C0 A0A2Y9DFV9 B7PRX4 A0A091S5I5 I3MMP7 A0A2K6BLP5 G7PJE1 A0A2K5VNY9 A0A0D9S2H4 A0A096MN14 A0A2K5N1T9 A0A091I6Q2 A0A2Y9IY17 A0A2J8MZR6 A0A131XDK8
Pubmed
26354079
22118469
26227816
24845553
28812685
20360741
+ More
23258410 19892987 19393038 22751099 15562597 15592404 20809919 17975172 21993624 28797301 25243066 26830274 23254933 20010809 20838655 16341006 21709235 16136131 18563158 21993625 22722832 30282656 22002653 25319552 25576852 10470851 14702039 16541075 15489334 19922875 20498093 20068231 21498477 21406692 23186163 23676666 21269460 26358130 28049606
23258410 19892987 19393038 22751099 15562597 15592404 20809919 17975172 21993624 28797301 25243066 26830274 23254933 20010809 20838655 16341006 21709235 16136131 18563158 21993625 22722832 30282656 22002653 25319552 25576852 10470851 14702039 16541075 15489334 19922875 20498093 20068231 21498477 21406692 23186163 23676666 21269460 26358130 28049606
EMBL
ODYU01002048
SOQ39070.1
NWSH01000510
PCG75954.1
GDQN01010696
JAT80358.1
+ More
RSAL01000020 RVE52649.1 KQ459603 KPI92659.1 KQ460845 KPJ12062.1 AGBW02012510 OWR44958.1 JTDY01006866 KOB65634.1 GDQN01003722 JAT87332.1 KK852819 KDR15615.1 NEVH01020852 PNF21205.1 NEDP02003393 OWF48767.1 PNF21207.1 JH431832 ABQF01016176 ABQF01016177 ABQF01016178 ABQF01016179 KB030754 ELK10309.1 GACC01000440 JAA74144.1 JH881492 ELR54014.1 AEMK02000037 AAQR03016528 AAQR03016529 AAQR03016530 AC145942 LSYS01004200 OPJ80451.1 KL225530 KFW73469.1 AMGL01082029 AMGL01082030 AANG04003655 AAPE02025061 AAPE02025062 GFPF01005446 MAA16592.1 KK686367 KFQ14733.1 KK649298 KFQ00844.1 GAMT01007944 GAMT01007943 GAMS01001101 GAMS01001100 GAMR01004756 GAMQ01003256 GAMQ01003255 GAMP01005983 GAMP01005982 JAB03917.1 JAB22035.1 JAB29176.1 JAB38595.1 JAB46772.1 GEDV01002006 JAP86551.1 AMQN01006786 KB299138 ELU08313.1 AGTO01000072 KL505791 KFO84794.1 GL192434 EFB13462.1 KK432927 KFQ89455.1 KK719069 KFO61211.1 KL289504 KFZ69370.1 AAEX03007381 KL447761 KFO76969.1 AEFK01192082 AEFK01192083 AEFK01192084 AACZ04070390 GABC01004182 GABD01004389 GABE01000614 NBAG03000240 JAA07156.1 JAA28711.1 JAA44125.1 PNI65014.1 GEFM01003926 JAP71870.1 GG666553 EEN56084.1 JH173279 EHB16325.1 AJFE02117077 AJFE02117078 AJFE02117079 AJFE02117080 NDHI03003364 PNJ83062.1 AEYP01012424 QUSF01000028 RLW00070.1 AAKN02055453 CM001263 EHH21135.1 JU320457 JU471911 AFE64213.1 AFH28715.1 GACK01005336 JAA59698.1 KL335586 KFZ49614.1 AB028956 AK001657 BC031358 AC016257 BC040936 BC104992 BC104994 BC110850 KL475304 KFV20369.1 GDAY02001210 JAV50217.1 KK707071 KFQ31010.1 ABJB010196317 DS775456 EEC09346.1 KK941274 KFQ50881.1 AGTP01024622 CM001286 EHH66661.1 AQIA01012545 AQIA01012546 AQIB01024467 AQIB01024468 AHZZ02002901 AHZZ02002902 KL217466 KFO95425.1 PNI65016.1 GEFH01004346 JAP64235.1
RSAL01000020 RVE52649.1 KQ459603 KPI92659.1 KQ460845 KPJ12062.1 AGBW02012510 OWR44958.1 JTDY01006866 KOB65634.1 GDQN01003722 JAT87332.1 KK852819 KDR15615.1 NEVH01020852 PNF21205.1 NEDP02003393 OWF48767.1 PNF21207.1 JH431832 ABQF01016176 ABQF01016177 ABQF01016178 ABQF01016179 KB030754 ELK10309.1 GACC01000440 JAA74144.1 JH881492 ELR54014.1 AEMK02000037 AAQR03016528 AAQR03016529 AAQR03016530 AC145942 LSYS01004200 OPJ80451.1 KL225530 KFW73469.1 AMGL01082029 AMGL01082030 AANG04003655 AAPE02025061 AAPE02025062 GFPF01005446 MAA16592.1 KK686367 KFQ14733.1 KK649298 KFQ00844.1 GAMT01007944 GAMT01007943 GAMS01001101 GAMS01001100 GAMR01004756 GAMQ01003256 GAMQ01003255 GAMP01005983 GAMP01005982 JAB03917.1 JAB22035.1 JAB29176.1 JAB38595.1 JAB46772.1 GEDV01002006 JAP86551.1 AMQN01006786 KB299138 ELU08313.1 AGTO01000072 KL505791 KFO84794.1 GL192434 EFB13462.1 KK432927 KFQ89455.1 KK719069 KFO61211.1 KL289504 KFZ69370.1 AAEX03007381 KL447761 KFO76969.1 AEFK01192082 AEFK01192083 AEFK01192084 AACZ04070390 GABC01004182 GABD01004389 GABE01000614 NBAG03000240 JAA07156.1 JAA28711.1 JAA44125.1 PNI65014.1 GEFM01003926 JAP71870.1 GG666553 EEN56084.1 JH173279 EHB16325.1 AJFE02117077 AJFE02117078 AJFE02117079 AJFE02117080 NDHI03003364 PNJ83062.1 AEYP01012424 QUSF01000028 RLW00070.1 AAKN02055453 CM001263 EHH21135.1 JU320457 JU471911 AFE64213.1 AFH28715.1 GACK01005336 JAA59698.1 KL335586 KFZ49614.1 AB028956 AK001657 BC031358 AC016257 BC040936 BC104992 BC104994 BC110850 KL475304 KFV20369.1 GDAY02001210 JAV50217.1 KK707071 KFQ31010.1 ABJB010196317 DS775456 EEC09346.1 KK941274 KFQ50881.1 AGTP01024622 CM001286 EHH66661.1 AQIA01012545 AQIA01012546 AQIB01024467 AQIB01024468 AHZZ02002901 AHZZ02002902 KL217466 KFO95425.1 PNI65016.1 GEFH01004346 JAP64235.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000027135 UP000235965 UP000242188 UP000085678 UP000007754 UP000010552 UP000002281 UP000009136 UP000008227 UP000005225 UP000076420 UP000000539 UP000190648 UP000054081 UP000002356 UP000011712 UP000001074 UP000252040 UP000245320 UP000248483 UP000014760 UP000016665 UP000008225 UP000286642 UP000079721 UP000245340 UP000261681 UP000233020 UP000233220 UP000001645 UP000286641 UP000233040 UP000248481 UP000052976 UP000002254 UP000248484 UP000248482 UP000053760 UP000007648 UP000286640 UP000002277 UP000001554 UP000006813 UP000240080 UP000000715 UP000276834 UP000005447 UP000261680 UP000005640 UP000233060 UP000248480 UP000001555 UP000005215 UP000233120 UP000009130 UP000233100 UP000029965 UP000028761 UP000054308
UP000027135 UP000235965 UP000242188 UP000085678 UP000007754 UP000010552 UP000002281 UP000009136 UP000008227 UP000005225 UP000076420 UP000000539 UP000190648 UP000054081 UP000002356 UP000011712 UP000001074 UP000252040 UP000245320 UP000248483 UP000014760 UP000016665 UP000008225 UP000286642 UP000079721 UP000245340 UP000261681 UP000233020 UP000233220 UP000001645 UP000286641 UP000233040 UP000248481 UP000052976 UP000002254 UP000248484 UP000248482 UP000053760 UP000007648 UP000286640 UP000002277 UP000001554 UP000006813 UP000240080 UP000000715 UP000276834 UP000005447 UP000261680 UP000005640 UP000233060 UP000248480 UP000001555 UP000005215 UP000233120 UP000009130 UP000233100 UP000029965 UP000028761 UP000054308
ProteinModelPortal
A0A2H1VDY1
A0A2A4JWE3
A0A1E1W053
A0A3S2LFI3
A0A194PIW6
A0A194R2Z4
+ More
A0A212ETZ5 A0A0L7KRI2 A0A1E1WK11 A0A067QYP0 A0A2J7PXZ5 A0A210QJ49 A0A2J7PXZ8 T1J3X6 A0A1S3JFP0 H0ZLG1 L5KF43 M3VK14 A0A3Q2IAN7 G5E600 L8IEP9 I3LLV0 H0WQT5 A0A2C9JNT2 R4GHH2 A0A1V4K7W3 A0A093RRJ7 W5Q6S3 M3VWP4 G1P956 A0A341AB65 A0A2U3ZZ47 A0A2Y9QKE1 A0A224YRZ8 A0A091Q5C7 A0A091P062 U3EJC9 A0A131Z8M7 R7UQC0 U3KG72 F7HA23 A0A3Q7WYU4 A0A091GNN6 D2H1T9 A0A1S3ACV0 A0A2U3WXU5 A0A383YSH6 A0A091UID3 A0A2K5F184 A0A2K6UPC6 G1NJT1 A0A3Q7PT10 A0A2K5RWX0 A0A2Y9H0S5 A0A3Q7Q7T4 A0A091EW44 A0A094LF41 J9P227 A0A2Y9T2K3 A0A2Y9IU07 A0A2C9JNS4 A0A2C9JNS6 A0A091G738 G3W5X6 A0A3Q7TN06 H2R755 A0A131Y0E5 C3YUG4 G5C464 A0A2R9CAM4 A0A2J8XM21 A0A2Y9IRC4 M3YL02 A0A3L8SCM6 H0V7S3 G7N5B6 H9EPA4 L7M974 A0A094K1P4 A0A384DAB7 A0A2U3ZUV1 Q2M389 A0A093D5J2 A0A1W7RGE6 A0A091QTV7 A0A2K5N2C0 A0A2Y9DFV9 B7PRX4 A0A091S5I5 I3MMP7 A0A2K6BLP5 G7PJE1 A0A2K5VNY9 A0A0D9S2H4 A0A096MN14 A0A2K5N1T9 A0A091I6Q2 A0A2Y9IY17 A0A2J8MZR6 A0A131XDK8
A0A212ETZ5 A0A0L7KRI2 A0A1E1WK11 A0A067QYP0 A0A2J7PXZ5 A0A210QJ49 A0A2J7PXZ8 T1J3X6 A0A1S3JFP0 H0ZLG1 L5KF43 M3VK14 A0A3Q2IAN7 G5E600 L8IEP9 I3LLV0 H0WQT5 A0A2C9JNT2 R4GHH2 A0A1V4K7W3 A0A093RRJ7 W5Q6S3 M3VWP4 G1P956 A0A341AB65 A0A2U3ZZ47 A0A2Y9QKE1 A0A224YRZ8 A0A091Q5C7 A0A091P062 U3EJC9 A0A131Z8M7 R7UQC0 U3KG72 F7HA23 A0A3Q7WYU4 A0A091GNN6 D2H1T9 A0A1S3ACV0 A0A2U3WXU5 A0A383YSH6 A0A091UID3 A0A2K5F184 A0A2K6UPC6 G1NJT1 A0A3Q7PT10 A0A2K5RWX0 A0A2Y9H0S5 A0A3Q7Q7T4 A0A091EW44 A0A094LF41 J9P227 A0A2Y9T2K3 A0A2Y9IU07 A0A2C9JNS4 A0A2C9JNS6 A0A091G738 G3W5X6 A0A3Q7TN06 H2R755 A0A131Y0E5 C3YUG4 G5C464 A0A2R9CAM4 A0A2J8XM21 A0A2Y9IRC4 M3YL02 A0A3L8SCM6 H0V7S3 G7N5B6 H9EPA4 L7M974 A0A094K1P4 A0A384DAB7 A0A2U3ZUV1 Q2M389 A0A093D5J2 A0A1W7RGE6 A0A091QTV7 A0A2K5N2C0 A0A2Y9DFV9 B7PRX4 A0A091S5I5 I3MMP7 A0A2K6BLP5 G7PJE1 A0A2K5VNY9 A0A0D9S2H4 A0A096MN14 A0A2K5N1T9 A0A091I6Q2 A0A2Y9IY17 A0A2J8MZR6 A0A131XDK8
Ontologies
PANTHER
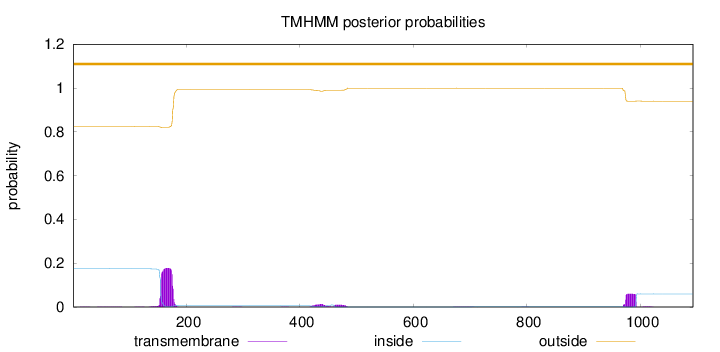
Topology
Subcellular location
Early endosome
Length:
1093
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.70244999999999
Exp number, first 60 AAs:
0.00965
Total prob of N-in:
0.17568
outside
1 - 1093
Population Genetic Test Statistics
Pi
1.659944
Theta
5.953028
Tajima's D
-2.094619
CLR
127.863098
CSRT
0.00789960501974901
Interpretation
Uncertain
