Pre Gene Modal
BGIBMGA010670
Annotation
PREDICTED:_pre-mRNA-processing_factor_40_homolog_A_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.098
Sequence
CDS
ATGGAGCCACCCAGTGCGGGGACCAGTTCTCCGGGTCTCTTAAACGCGCCTCCTCTTTTACCGCCTCCAATGCTAAGCGGTATGCCTCCACCAATGCCGCCCGCTGTGGCAATGCCACCAGTGCCTGGGTTACCTCCAAATATGGCTACATTACCACCGCCCATGGGCTTTCCTCCCATGTTGCCGCCTCCATTTTCAATACCACCGCCAGGATTCCCTGCATTCAAACCGGAATTAAATGCTCCTGCACCAGATGTTGCTCCATCGGCAAATCAAAGTAGTCCATGGTCTGAACATAAGGCCCCTGATGGAAGAACATATTACTATAATTCTGTAACCAAACAAAGCTTGTGGGAAAAACCTGACGACTTAAAAACGTCAGCTGAGAAACTATTATCTGCTTGTGTATGGAAAGAATATACCACAAATACTGGCAGACTTTACTATCATAATATTGAGACAAAGGAGTCCAGTTGGGTAGTACCGAAAGAATTACAGGAGATTAAAGACAAAATTGCTGCTGAAGAAGCTGCTCAGCGCATTGTGATGGAAGTAACACCGAGTGAGGTGCCGCTGCCGGCAGAGCCTGCAGGCTCCTCCGCCCTTGACGAAGCTATGGCTAAAACATTAGCAGCCATTGATACCTCATTGGCCACATCCATACCTGTACCTGACGAAAAACCAGAAGACACAGTTTCAATTAATAATGAAACTCAAGAACAACCTGAGATGTTGTACAAAGACAAGAAAGAAGCTATTGAAGCATTTAAAGAACTCCTGAAAGATAAGAATATACCCTCAAATGCAACATGGGAACAATGTGTAAAAGTTATTTCAAAAGATCCACGATATGTCACTTTTAAAAAACTCAATGAAAAGAAACAAGCGTTCAATGCTTACAAAACTCAAAAACTAAAAGATGAAAGAGAGGAACAGAGACTGAAAACAAAAAAAAATCGCGAGAATCTTGAAGAGTTTTTGTTGAGTTGCGAACGGGTGACATCTCTAACAAAATTTTACAAATGTGAAGAAATGTTCAGCAATCTAGAGATTTGGCGATGTGTTCCAGAAGCCGATCGTAGGGACATATTTGAAGATTGTATTTTTACTATAGCGAAACGGGAAAAAGAAGAAGCAAAGGCTTTAAAGAAACGGAACATGAAAATGTTAACCCAGGTATTAGAGAACATGAGCGAAATTACGTACAGTAGTACGTGGAGTGAGGCACAAGTACTATTGCTAGAAAATTCAGCTTTTAAAAATGATGTTAGTTTACTAGGTATGGACAAGGAAGATGCCCTGATTGTATTTGAACAACACATACGAAATTTAGAAGCTGAATATCAACAAGAAAGAGAACAAATTAAAAAGAGGAATAAAAGACAACAAAGAAAAAATAGAGACAACTTTTTGGCTTTGCTGGACAGTCTTCATGAAGAAGGTAAATTGACATCGATGTCTCTATGGGTCGAGCTGTATCCGGTTATTTCTGCAGACATGAGATTTTCAGCTATGTTAGGACAAAGCGGATCGACACCGTTAGACTTATTCAAGTTTTACGTTGAAAATTTGAAAGCGAGGTTCCACGACGAAAAGAAGATAATAAAGGAGATACTGAAAGACAAGAATTTCGAAGTTAAACCGGAGACCACGTTCGAGGAGTTCGCCACCGTCGTGTGCGAGGACAGCAAGTCCGCCTCGCTCGACGCCGGAAATGTCAAACTTACTTACAATTCGCTTTTAGAGAAGGCTGAAACTAGAAAGAAGGAGAAATTGAAAGAAGAATCGAAAGCTCAAAAGAAAATAGAATCTGCATTTAAATGGGCATTGAGTGATGCAAACATCGACCACCTGCTCTCGTGGAGCGAGGTTAAGGAGAAACTTGACTTGAACGCGCCAGAGTTTGCCGCCGTCACCAGTGAAGAAGACAGAATTAGAATTTATAAGGATTATCAGCATGAACAAGAAGAAAGCTGCATGCACTATCACCATCCAAAACCAAAGAAGTCCAAACGATCAAAGAAGAAGAAGCGTTCTCACTCTGCCTCTTTGTCTCGGTCAGCATCAGCGTCGCGGTCGCCATCCCCGGCACCGAGCCACGGCACTTGGAGCTCCGACGAGGGCCGGAAACCCAAGAAAACTAAGAAGAAGCATCGGAGGCACTCTCCTGCTCCGAAATCACCGTCTCCCGAGGAGGGTGGCATCACAGACGACGAAGAAGTGATTCGGCACAAAAACAAAAAGACGAAACGCAGTGCGCCCAGCAGCCCGGAAGAGGAACAGGAACCGGAAATCGTTTACAAACCCAAGAAGAAGAAGGACAAGAAAGATAAAAAAGAGAGGTCGGGTGCGGCGTGGAGTGACGCCGACCTGGAGTCGCGTCGTGCGGCGCTGCTAGCTCAGCTGCACGAGCACGAGGCGGACTGA
Protein
MEPPSAGTSSPGLLNAPPLLPPPMLSGMPPPMPPAVAMPPVPGLPPNMATLPPPMGFPPMLPPPFSIPPPGFPAFKPELNAPAPDVAPSANQSSPWSEHKAPDGRTYYYNSVTKQSLWEKPDDLKTSAEKLLSACVWKEYTTNTGRLYYHNIETKESSWVVPKELQEIKDKIAAEEAAQRIVMEVTPSEVPLPAEPAGSSALDEAMAKTLAAIDTSLATSIPVPDEKPEDTVSINNETQEQPEMLYKDKKEAIEAFKELLKDKNIPSNATWEQCVKVISKDPRYVTFKKLNEKKQAFNAYKTQKLKDEREEQRLKTKKNRENLEEFLLSCERVTSLTKFYKCEEMFSNLEIWRCVPEADRRDIFEDCIFTIAKREKEEAKALKKRNMKMLTQVLENMSEITYSSTWSEAQVLLLENSAFKNDVSLLGMDKEDALIVFEQHIRNLEAEYQQEREQIKKRNKRQQRKNRDNFLALLDSLHEEGKLTSMSLWVELYPVISADMRFSAMLGQSGSTPLDLFKFYVENLKARFHDEKKIIKEILKDKNFEVKPETTFEEFATVVCEDSKSASLDAGNVKLTYNSLLEKAETRKKEKLKEESKAQKKIESAFKWALSDANIDHLLSWSEVKEKLDLNAPEFAAVTSEEDRIRIYKDYQHEQEESCMHYHHPKPKKSKRSKKKKRSHSASLSRSASASRSPSPAPSHGTWSSDEGRKPKKTKKKHRRHSPAPKSPSPEEGGITDDEEVIRHKNKKTKRSAPSSPEEEQEPEIVYKPKKKKDKKDKKERSGAAWSDADLESRRAALLAQLHEHEAD
Summary
Uniprot
H9JMB7
A0A2A4JVB7
A0A2H1VE01
A0A194PHY2
A0A194R2P6
A0A1E1W155
+ More
D2A466 A0A1W4WVV7 A0A1Y1K8C7 A0A1Y1KG09 A0A1Y1K8G9 A0A0L7QNR5 A0A154PPP6 A0A088A1I7 A0A2A3EII4 A0A026X1E2 A0A1Y1KDJ1 A0A1Y1KB37 A0A1Y1K872 E2ACC7 A0A0C9RKZ6 F4WZV6 A0A0J7L715 A0A158NH65 E2BU72 A0A232F700 E9IL98 A0A151ICF1 A0A1Y1KE41 A0A1Y1K8F6 N6U607 K7IVK3 U4U4T0 A0A151XAH3 A0A195B719 A0A195FFP4 A0A195DXK7 A0A1B0D1J8 A0A1B0C8A0 A0A182MW53 A0A182Y3H6 A0A336LL88 A0A336LQS5 A0A336LQA8 A0A182R2Z8 A0A336LL83 A0A336LYI3 A0A182W6R1 Q17BB3 A0A182NU18 Q7PNE5 A0A182XL43 A0A1S4GXR9 A0A182ICH9 A0A182JY97 A0A182UVP3 A0A182TVT5 W5JFM9 U5ESU8 A0A084W4U2 A0A1B6EA38 A0A0P4WBT2 A0A1Q3FW24 A0A0N7ZBZ3 A0A1Q3F1D3 A0A182GKQ7 A0A182PCN5 A0A182GQ15 A0A0P4WEZ2 A0A0P4W9L4 A0A3Q0JD52 A0A1I8PA21 A0A1I8P9U2 A0A3Q0J859 A0A182UIZ9 A0A1S4EK48 B0X9W9 A0A0L0BZF9 E9HFR0 A0A1I8NAE5 B3MMY5 A0A0A9W9D7 A0A1B0G9K7 A0A1A9Z7U5 B4MW11 A0A1A9V864 A0A3B0JH08 A0A3B0K3F4 Q29LZ7 A0A1A9XEN0 A0A0R3P0A0 B4G6Z5 A0A1B0C570 A0A0J9QVI7 A0A0A9W9E5 A0A0A9WBA7 A0A0P5DV99 A0A0P6ACK2 Q9VQK5
D2A466 A0A1W4WVV7 A0A1Y1K8C7 A0A1Y1KG09 A0A1Y1K8G9 A0A0L7QNR5 A0A154PPP6 A0A088A1I7 A0A2A3EII4 A0A026X1E2 A0A1Y1KDJ1 A0A1Y1KB37 A0A1Y1K872 E2ACC7 A0A0C9RKZ6 F4WZV6 A0A0J7L715 A0A158NH65 E2BU72 A0A232F700 E9IL98 A0A151ICF1 A0A1Y1KE41 A0A1Y1K8F6 N6U607 K7IVK3 U4U4T0 A0A151XAH3 A0A195B719 A0A195FFP4 A0A195DXK7 A0A1B0D1J8 A0A1B0C8A0 A0A182MW53 A0A182Y3H6 A0A336LL88 A0A336LQS5 A0A336LQA8 A0A182R2Z8 A0A336LL83 A0A336LYI3 A0A182W6R1 Q17BB3 A0A182NU18 Q7PNE5 A0A182XL43 A0A1S4GXR9 A0A182ICH9 A0A182JY97 A0A182UVP3 A0A182TVT5 W5JFM9 U5ESU8 A0A084W4U2 A0A1B6EA38 A0A0P4WBT2 A0A1Q3FW24 A0A0N7ZBZ3 A0A1Q3F1D3 A0A182GKQ7 A0A182PCN5 A0A182GQ15 A0A0P4WEZ2 A0A0P4W9L4 A0A3Q0JD52 A0A1I8PA21 A0A1I8P9U2 A0A3Q0J859 A0A182UIZ9 A0A1S4EK48 B0X9W9 A0A0L0BZF9 E9HFR0 A0A1I8NAE5 B3MMY5 A0A0A9W9D7 A0A1B0G9K7 A0A1A9Z7U5 B4MW11 A0A1A9V864 A0A3B0JH08 A0A3B0K3F4 Q29LZ7 A0A1A9XEN0 A0A0R3P0A0 B4G6Z5 A0A1B0C570 A0A0J9QVI7 A0A0A9W9E5 A0A0A9WBA7 A0A0P5DV99 A0A0P6ACK2 Q9VQK5
Pubmed
19121390
26354079
18362917
19820115
28004739
24508170
+ More
20798317 21719571 21347285 28648823 21282665 23537049 20075255 25244985 17510324 12364791 20920257 23761445 24438588 26483478 26108605 21292972 25315136 17994087 25401762 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
20798317 21719571 21347285 28648823 21282665 23537049 20075255 25244985 17510324 12364791 20920257 23761445 24438588 26483478 26108605 21292972 25315136 17994087 25401762 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01033439
BABH01033440
BABH01033441
NWSH01000510
PCG75951.1
ODYU01002047
+ More
SOQ39068.1 KQ459603 KPI92658.1 KQ460845 KPJ12063.1 GDQN01010348 JAT80706.1 KQ971348 EFA05623.1 GEZM01089113 JAV57689.1 GEZM01089115 JAV57687.1 GEZM01089118 JAV57684.1 KQ414851 KOC60209.1 KQ435012 KZC13866.1 KZ288241 PBC31284.1 KK107063 EZA61199.1 GEZM01089117 JAV57685.1 GEZM01089114 JAV57688.1 GEZM01089120 JAV57682.1 GL438491 EFN68917.1 GBYB01007586 JAG77353.1 GL888480 EGI60246.1 LBMM01000397 KMQ98458.1 ADTU01015146 GL450586 EFN80756.1 NNAY01000799 OXU26445.1 GL764074 EFZ18647.1 KQ978053 KYM97764.1 GEZM01089116 JAV57686.1 GEZM01089119 JAV57683.1 APGK01047620 APGK01047621 APGK01047622 KB741084 ENN74007.1 KB632108 ERL88889.1 KQ982339 KYQ57357.1 KQ976574 KYM80313.1 KQ981625 KYN39047.1 KQ980107 KYN17613.1 AJVK01022090 AJWK01000296 AXCM01001956 AXCM01001957 UFQT01000042 SSX18726.1 SSX18727.1 SSX18729.1 SSX18730.1 SSX18728.1 CH477324 EAT43533.1 AAAB01008964 EAA12318.5 APCN01000799 ADMH02001645 ETN61619.1 GANO01003066 JAB56805.1 ATLV01020383 KE525299 KFB45236.1 GEDC01002502 JAS34796.1 GDRN01075703 JAI63012.1 GFDL01003224 JAV31821.1 GDRN01075709 GDRN01075708 JAI63008.1 GFDL01013679 JAV21366.1 JXUM01070412 KQ562604 KXJ75509.1 JXUM01079561 KQ563134 KXJ74426.1 GDRN01075707 JAI63009.1 GDRN01075706 JAI63010.1 DS232553 EDS43395.1 JRES01001205 KNC24629.1 GL732637 EFX69363.1 CH902620 EDV31010.1 GBHO01038552 GBRD01007343 JAG05052.1 JAG58478.1 CCAG010009121 CH963857 EDW75881.1 OUUW01000004 SPP79562.1 SPP79561.1 CH379060 EAL33899.1 KRT04310.1 CH479180 EDW29194.1 JXJN01010999 JXJN01025882 CM002910 KMY87804.1 GBHO01038542 JAG05062.1 GBHO01038540 JAG05064.1 GDIP01157075 JAJ66327.1 GDIP01036250 JAM67465.1 AE014134 AY058551 AAF51160.1 AAL13780.1
SOQ39068.1 KQ459603 KPI92658.1 KQ460845 KPJ12063.1 GDQN01010348 JAT80706.1 KQ971348 EFA05623.1 GEZM01089113 JAV57689.1 GEZM01089115 JAV57687.1 GEZM01089118 JAV57684.1 KQ414851 KOC60209.1 KQ435012 KZC13866.1 KZ288241 PBC31284.1 KK107063 EZA61199.1 GEZM01089117 JAV57685.1 GEZM01089114 JAV57688.1 GEZM01089120 JAV57682.1 GL438491 EFN68917.1 GBYB01007586 JAG77353.1 GL888480 EGI60246.1 LBMM01000397 KMQ98458.1 ADTU01015146 GL450586 EFN80756.1 NNAY01000799 OXU26445.1 GL764074 EFZ18647.1 KQ978053 KYM97764.1 GEZM01089116 JAV57686.1 GEZM01089119 JAV57683.1 APGK01047620 APGK01047621 APGK01047622 KB741084 ENN74007.1 KB632108 ERL88889.1 KQ982339 KYQ57357.1 KQ976574 KYM80313.1 KQ981625 KYN39047.1 KQ980107 KYN17613.1 AJVK01022090 AJWK01000296 AXCM01001956 AXCM01001957 UFQT01000042 SSX18726.1 SSX18727.1 SSX18729.1 SSX18730.1 SSX18728.1 CH477324 EAT43533.1 AAAB01008964 EAA12318.5 APCN01000799 ADMH02001645 ETN61619.1 GANO01003066 JAB56805.1 ATLV01020383 KE525299 KFB45236.1 GEDC01002502 JAS34796.1 GDRN01075703 JAI63012.1 GFDL01003224 JAV31821.1 GDRN01075709 GDRN01075708 JAI63008.1 GFDL01013679 JAV21366.1 JXUM01070412 KQ562604 KXJ75509.1 JXUM01079561 KQ563134 KXJ74426.1 GDRN01075707 JAI63009.1 GDRN01075706 JAI63010.1 DS232553 EDS43395.1 JRES01001205 KNC24629.1 GL732637 EFX69363.1 CH902620 EDV31010.1 GBHO01038552 GBRD01007343 JAG05052.1 JAG58478.1 CCAG010009121 CH963857 EDW75881.1 OUUW01000004 SPP79562.1 SPP79561.1 CH379060 EAL33899.1 KRT04310.1 CH479180 EDW29194.1 JXJN01010999 JXJN01025882 CM002910 KMY87804.1 GBHO01038542 JAG05062.1 GBHO01038540 JAG05064.1 GDIP01157075 JAJ66327.1 GDIP01036250 JAM67465.1 AE014134 AY058551 AAF51160.1 AAL13780.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007266
UP000192223
+ More
UP000053825 UP000076502 UP000005203 UP000242457 UP000053097 UP000000311 UP000007755 UP000036403 UP000005205 UP000008237 UP000215335 UP000078542 UP000019118 UP000002358 UP000030742 UP000075809 UP000078540 UP000078541 UP000078492 UP000092462 UP000092461 UP000075883 UP000076408 UP000075900 UP000075920 UP000008820 UP000075884 UP000007062 UP000076407 UP000075840 UP000075881 UP000075903 UP000075902 UP000000673 UP000030765 UP000069940 UP000249989 UP000075885 UP000079169 UP000095300 UP000002320 UP000037069 UP000000305 UP000095301 UP000007801 UP000092444 UP000092445 UP000007798 UP000078200 UP000268350 UP000001819 UP000092443 UP000008744 UP000092460 UP000000803
UP000053825 UP000076502 UP000005203 UP000242457 UP000053097 UP000000311 UP000007755 UP000036403 UP000005205 UP000008237 UP000215335 UP000078542 UP000019118 UP000002358 UP000030742 UP000075809 UP000078540 UP000078541 UP000078492 UP000092462 UP000092461 UP000075883 UP000076408 UP000075900 UP000075920 UP000008820 UP000075884 UP000007062 UP000076407 UP000075840 UP000075881 UP000075903 UP000075902 UP000000673 UP000030765 UP000069940 UP000249989 UP000075885 UP000079169 UP000095300 UP000002320 UP000037069 UP000000305 UP000095301 UP000007801 UP000092444 UP000092445 UP000007798 UP000078200 UP000268350 UP000001819 UP000092443 UP000008744 UP000092460 UP000000803
Pfam
Interpro
IPR036517
FF_domain_sf
+ More
IPR002713 FF_domain
IPR001202 WW_dom
IPR036020 WW_dom_sf
IPR008521 Mg_trans_NIPA
IPR039726 Prp40-like
IPR016161 Ald_DH/histidinol_DH
IPR006638 Elp3/MiaB/NifB
IPR016160 Ald_DH_CS_CYS
IPR016163 Ald_DH_C
IPR006463 MiaB_methiolase
IPR005839 Methylthiotransferase
IPR016162 Ald_DH_N
IPR005931 P5CDH/ALDH4A1
IPR023404 rSAM_horseshoe
IPR013848 Methylthiotransferase_N
IPR020612 Methylthiotransferase_CS
IPR038135 Methylthiotransferase_N_sf
IPR015590 Aldehyde_DH_dom
IPR007197 rSAM
IPR002792 TRAM_dom
IPR018247 EF_Hand_1_Ca_BS
IPR003124 WH2_dom
IPR035979 RBD_domain_sf
IPR036443 Znf_RanBP2_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR001876 Znf_RanBP2
IPR000504 RRM_dom
IPR002713 FF_domain
IPR001202 WW_dom
IPR036020 WW_dom_sf
IPR008521 Mg_trans_NIPA
IPR039726 Prp40-like
IPR016161 Ald_DH/histidinol_DH
IPR006638 Elp3/MiaB/NifB
IPR016160 Ald_DH_CS_CYS
IPR016163 Ald_DH_C
IPR006463 MiaB_methiolase
IPR005839 Methylthiotransferase
IPR016162 Ald_DH_N
IPR005931 P5CDH/ALDH4A1
IPR023404 rSAM_horseshoe
IPR013848 Methylthiotransferase_N
IPR020612 Methylthiotransferase_CS
IPR038135 Methylthiotransferase_N_sf
IPR015590 Aldehyde_DH_dom
IPR007197 rSAM
IPR002792 TRAM_dom
IPR018247 EF_Hand_1_Ca_BS
IPR003124 WH2_dom
IPR035979 RBD_domain_sf
IPR036443 Znf_RanBP2_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR001876 Znf_RanBP2
IPR000504 RRM_dom
SUPFAM
ProteinModelPortal
H9JMB7
A0A2A4JVB7
A0A2H1VE01
A0A194PHY2
A0A194R2P6
A0A1E1W155
+ More
D2A466 A0A1W4WVV7 A0A1Y1K8C7 A0A1Y1KG09 A0A1Y1K8G9 A0A0L7QNR5 A0A154PPP6 A0A088A1I7 A0A2A3EII4 A0A026X1E2 A0A1Y1KDJ1 A0A1Y1KB37 A0A1Y1K872 E2ACC7 A0A0C9RKZ6 F4WZV6 A0A0J7L715 A0A158NH65 E2BU72 A0A232F700 E9IL98 A0A151ICF1 A0A1Y1KE41 A0A1Y1K8F6 N6U607 K7IVK3 U4U4T0 A0A151XAH3 A0A195B719 A0A195FFP4 A0A195DXK7 A0A1B0D1J8 A0A1B0C8A0 A0A182MW53 A0A182Y3H6 A0A336LL88 A0A336LQS5 A0A336LQA8 A0A182R2Z8 A0A336LL83 A0A336LYI3 A0A182W6R1 Q17BB3 A0A182NU18 Q7PNE5 A0A182XL43 A0A1S4GXR9 A0A182ICH9 A0A182JY97 A0A182UVP3 A0A182TVT5 W5JFM9 U5ESU8 A0A084W4U2 A0A1B6EA38 A0A0P4WBT2 A0A1Q3FW24 A0A0N7ZBZ3 A0A1Q3F1D3 A0A182GKQ7 A0A182PCN5 A0A182GQ15 A0A0P4WEZ2 A0A0P4W9L4 A0A3Q0JD52 A0A1I8PA21 A0A1I8P9U2 A0A3Q0J859 A0A182UIZ9 A0A1S4EK48 B0X9W9 A0A0L0BZF9 E9HFR0 A0A1I8NAE5 B3MMY5 A0A0A9W9D7 A0A1B0G9K7 A0A1A9Z7U5 B4MW11 A0A1A9V864 A0A3B0JH08 A0A3B0K3F4 Q29LZ7 A0A1A9XEN0 A0A0R3P0A0 B4G6Z5 A0A1B0C570 A0A0J9QVI7 A0A0A9W9E5 A0A0A9WBA7 A0A0P5DV99 A0A0P6ACK2 Q9VQK5
D2A466 A0A1W4WVV7 A0A1Y1K8C7 A0A1Y1KG09 A0A1Y1K8G9 A0A0L7QNR5 A0A154PPP6 A0A088A1I7 A0A2A3EII4 A0A026X1E2 A0A1Y1KDJ1 A0A1Y1KB37 A0A1Y1K872 E2ACC7 A0A0C9RKZ6 F4WZV6 A0A0J7L715 A0A158NH65 E2BU72 A0A232F700 E9IL98 A0A151ICF1 A0A1Y1KE41 A0A1Y1K8F6 N6U607 K7IVK3 U4U4T0 A0A151XAH3 A0A195B719 A0A195FFP4 A0A195DXK7 A0A1B0D1J8 A0A1B0C8A0 A0A182MW53 A0A182Y3H6 A0A336LL88 A0A336LQS5 A0A336LQA8 A0A182R2Z8 A0A336LL83 A0A336LYI3 A0A182W6R1 Q17BB3 A0A182NU18 Q7PNE5 A0A182XL43 A0A1S4GXR9 A0A182ICH9 A0A182JY97 A0A182UVP3 A0A182TVT5 W5JFM9 U5ESU8 A0A084W4U2 A0A1B6EA38 A0A0P4WBT2 A0A1Q3FW24 A0A0N7ZBZ3 A0A1Q3F1D3 A0A182GKQ7 A0A182PCN5 A0A182GQ15 A0A0P4WEZ2 A0A0P4W9L4 A0A3Q0JD52 A0A1I8PA21 A0A1I8P9U2 A0A3Q0J859 A0A182UIZ9 A0A1S4EK48 B0X9W9 A0A0L0BZF9 E9HFR0 A0A1I8NAE5 B3MMY5 A0A0A9W9D7 A0A1B0G9K7 A0A1A9Z7U5 B4MW11 A0A1A9V864 A0A3B0JH08 A0A3B0K3F4 Q29LZ7 A0A1A9XEN0 A0A0R3P0A0 B4G6Z5 A0A1B0C570 A0A0J9QVI7 A0A0A9W9E5 A0A0A9WBA7 A0A0P5DV99 A0A0P6ACK2 Q9VQK5
PDB
2L5F
E-value=1.71428e-21,
Score=256
Ontologies
KEGG
GO
PANTHER
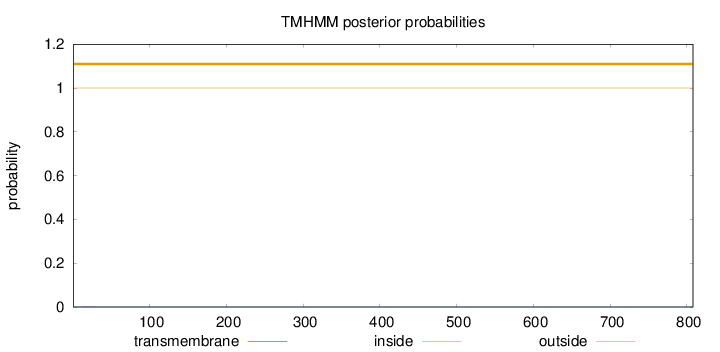
Topology
Length:
808
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00407999999999999
Exp number, first 60 AAs:
0.00407999999999999
Total prob of N-in:
0.00021
outside
1 - 808
Population Genetic Test Statistics
Pi
0.90556
Theta
4.377904
Tajima's D
-2.199996
CLR
303.073677
CSRT
0.00379981000949953
Interpretation
Possibly Positive selection
