Pre Gene Modal
BGIBMGA010670
Annotation
PREDICTED:_magnesium_transporter_NIPA2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.643
Sequence
CDS
ATGTGGTGGCTGGGATTAATAACAATGAGCATTGGTGAAGCAGCTAATTTGTTAGCATATGCATTTGCTCCAGCTGCATTAGTAACTCCACTTGGTGCCCTGAGTGTCCTGGTAGCTGCTGTGCTTTCTTCAAAGTTTTTAAATGAGAAGTTGAATTTTATGGGGAAGATTGCCTGTTTTCTATGTATTATTGGATCAATTGTATTTGTTATTCATTCTCCGAAATCAGAAGAAATTAAAAACTTTTCGGAAGTTGTGAATATGCTATAAGACACGTAA
Protein
MWWLGLITMSIGEAANLLAYAFAPAALVTPLGALSVLVAAVLSSKFLNEKLNFMGKIACFLCIIGSIVFVIHSPKSEEIKNFSEVVNMLXDT
Summary
Uniprot
A0A2A4J158
A0A194R3G6
A0A194PNF7
A0A2H1VE26
A0A3S2N3P1
S4PX69
+ More
A0A151WU50 D6X0E4 B4MV91 A0A212EU15 B4Q490 A0A0J9R1G7 Q9VK62 B3N3C9 M9PD12 B4IEC0 A0A1W4UT61 A0A182YDJ4 A0A0L7RDP4 B4JQD7 A0A1B6FJ15 B4P2I6 A0A195AZ37 A0A1B0CKY0 B4HB27 A0A158NYD7 B3MNJ0 A0A3B0JIB4 A0A182M406 A0A310SP55 B4MDM3 A0A154PB38 Q29L74 A0A151J5D7 A0A182NC11 E9JD25 A0A0M8ZYI2 A0A0A1XQX0 B4KIN1 A0A182T0N9 A0A336N1L2 A0A182JBC5 W8BRX5 A0A034WQ74 F4WSY5 A0A1A9VMF4 A0A1B0AEN7 D3TMV7 A0A0T6BF60 A0A1B6ML95 A0A195F7H0 A0A182QRT6 A0A1A9Y3K0 A0A1B0BLA1 A0A1A9WM35 A0A0L7KRJ4 A0A182WGR3 A0A182RBJ4 E2BE87 A0A026WBW2 Q7Q1D5 T1PGV5 A0A182JUG2 A0A182PLM8 A0A1I8M2M9 A0A182HRN8 A0A182V3J9 A0A182X346 A0A182KTS3 A0A182UBR9 A0A2A3EE13 A0A088AN02 A0A2S2QZ46 A0A0L0C6A6 A0A1I8CKL1 A0A0N4XN42 A0A1L8EI08 A0A1I8PRC3 E2AQJ9 A0A084W8U3 A0A2H8TK96 A0A1I7ZKV4 A0A2I4AUU5 A0A3P7YAY7 U5ES80 A0A146QY91 A0A2G9TQX1 A0A147AN85 T1DK97 A0A183GXW8 A0A0J7K383 A0A3Q2PJM6 T1E7M5 A0A3P8S3P4 A0A3Q3BBT7 A0A3P8S1D6 A0A1I8BLD1 A0A2M4APJ6 A0A2M4API0 A0A2M4APM3
A0A151WU50 D6X0E4 B4MV91 A0A212EU15 B4Q490 A0A0J9R1G7 Q9VK62 B3N3C9 M9PD12 B4IEC0 A0A1W4UT61 A0A182YDJ4 A0A0L7RDP4 B4JQD7 A0A1B6FJ15 B4P2I6 A0A195AZ37 A0A1B0CKY0 B4HB27 A0A158NYD7 B3MNJ0 A0A3B0JIB4 A0A182M406 A0A310SP55 B4MDM3 A0A154PB38 Q29L74 A0A151J5D7 A0A182NC11 E9JD25 A0A0M8ZYI2 A0A0A1XQX0 B4KIN1 A0A182T0N9 A0A336N1L2 A0A182JBC5 W8BRX5 A0A034WQ74 F4WSY5 A0A1A9VMF4 A0A1B0AEN7 D3TMV7 A0A0T6BF60 A0A1B6ML95 A0A195F7H0 A0A182QRT6 A0A1A9Y3K0 A0A1B0BLA1 A0A1A9WM35 A0A0L7KRJ4 A0A182WGR3 A0A182RBJ4 E2BE87 A0A026WBW2 Q7Q1D5 T1PGV5 A0A182JUG2 A0A182PLM8 A0A1I8M2M9 A0A182HRN8 A0A182V3J9 A0A182X346 A0A182KTS3 A0A182UBR9 A0A2A3EE13 A0A088AN02 A0A2S2QZ46 A0A0L0C6A6 A0A1I8CKL1 A0A0N4XN42 A0A1L8EI08 A0A1I8PRC3 E2AQJ9 A0A084W8U3 A0A2H8TK96 A0A1I7ZKV4 A0A2I4AUU5 A0A3P7YAY7 U5ES80 A0A146QY91 A0A2G9TQX1 A0A147AN85 T1DK97 A0A183GXW8 A0A0J7K383 A0A3Q2PJM6 T1E7M5 A0A3P8S3P4 A0A3Q3BBT7 A0A3P8S1D6 A0A1I8BLD1 A0A2M4APJ6 A0A2M4API0 A0A2M4APM3
Pubmed
26354079
23622113
18362917
19820115
17994087
22118469
+ More
22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 17550304 21347285 15632085 21282665 25830018 24495485 25348373 21719571 20353571 26227816 20798317 24508170 12364791 14747013 17210077 25315136 20966253 26108605 26829753 24438588 26392177 18809916
22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 17550304 21347285 15632085 21282665 25830018 24495485 25348373 21719571 20353571 26227816 20798317 24508170 12364791 14747013 17210077 25315136 20966253 26108605 26829753 24438588 26392177 18809916
EMBL
NWSH01004441
PCG65113.1
KQ460845
KPJ12064.1
KQ459603
KPI92655.1
+ More
ODYU01002047 SOQ39067.1 RSAL01002015 RVE40604.1 GAIX01005558 JAA87002.1 KQ982750 KYQ51271.1 KQ971372 EFA09585.1 CH963857 EDW76436.1 AGBW02012509 OWR44969.1 CM000361 EDX04840.1 CM002910 KMY89956.1 AE014134 AY089521 AAF53217.1 AAL90259.1 CH954177 EDV58769.2 AGB92956.1 CH480831 EDW45947.1 KQ414613 KOC68939.1 CH916372 EDV99117.1 GECZ01024225 GECZ01019578 GECZ01002712 GECZ01001315 JAS45544.1 JAS50191.1 JAS67057.1 JAS68454.1 CM000157 EDW88216.2 KQ976701 KYM77219.1 AJWK01016765 CH479247 EDW37830.1 ADTU01003763 CH902620 EDV32098.2 OUUW01000006 SPP82097.1 AXCM01001931 KQ761768 OAD57112.1 CH940661 EDW71284.2 KQ434864 KZC09053.1 CH379061 EAL32950.3 KQ980055 KYN17974.1 GL771866 EFZ09143.1 KQ435824 KOX72203.1 GBXI01001324 JAD12968.1 CH933807 EDW11374.2 UFQT01003132 SSX34617.1 GAMC01002445 JAC04111.1 GAKP01001241 GAKP01001240 JAC57711.1 GL888331 EGI62689.1 CCAG010021443 EZ422759 ADD19035.1 LJIG01000966 KRT85974.1 GEBQ01003328 JAT36649.1 KQ981768 KYN36014.1 AXCN02000699 JXJN01016272 JXJN01016273 JTDY01006531 KOB65927.1 GL447768 EFN85932.1 KK107295 EZA53106.1 AAAB01008980 EAA14313.3 KA648027 AFP62656.1 APCN01001728 KZ288269 PBC30023.1 GGMS01013199 MBY82402.1 JRES01000841 KNC27815.1 UYSL01006581 VDL67534.1 GFDG01000452 JAV18347.1 GL441773 EFN64308.1 ATLV01021515 KE525319 KFB46637.1 GFXV01002749 MBW14554.1 UZAH01025668 VDO68344.1 GANO01003365 JAB56506.1 GCES01125253 JAQ61069.1 KZ355594 PIO60384.1 GCES01006432 JAR79891.1 GAMD01001056 JAB00535.1 UZAJ01000018 VDO24431.1 LBMM01015885 KMQ84631.1 GAMD01002902 JAA98688.1 GGFK01009372 MBW42693.1 GGFK01009359 MBW42680.1 GGFK01009386 MBW42707.1
ODYU01002047 SOQ39067.1 RSAL01002015 RVE40604.1 GAIX01005558 JAA87002.1 KQ982750 KYQ51271.1 KQ971372 EFA09585.1 CH963857 EDW76436.1 AGBW02012509 OWR44969.1 CM000361 EDX04840.1 CM002910 KMY89956.1 AE014134 AY089521 AAF53217.1 AAL90259.1 CH954177 EDV58769.2 AGB92956.1 CH480831 EDW45947.1 KQ414613 KOC68939.1 CH916372 EDV99117.1 GECZ01024225 GECZ01019578 GECZ01002712 GECZ01001315 JAS45544.1 JAS50191.1 JAS67057.1 JAS68454.1 CM000157 EDW88216.2 KQ976701 KYM77219.1 AJWK01016765 CH479247 EDW37830.1 ADTU01003763 CH902620 EDV32098.2 OUUW01000006 SPP82097.1 AXCM01001931 KQ761768 OAD57112.1 CH940661 EDW71284.2 KQ434864 KZC09053.1 CH379061 EAL32950.3 KQ980055 KYN17974.1 GL771866 EFZ09143.1 KQ435824 KOX72203.1 GBXI01001324 JAD12968.1 CH933807 EDW11374.2 UFQT01003132 SSX34617.1 GAMC01002445 JAC04111.1 GAKP01001241 GAKP01001240 JAC57711.1 GL888331 EGI62689.1 CCAG010021443 EZ422759 ADD19035.1 LJIG01000966 KRT85974.1 GEBQ01003328 JAT36649.1 KQ981768 KYN36014.1 AXCN02000699 JXJN01016272 JXJN01016273 JTDY01006531 KOB65927.1 GL447768 EFN85932.1 KK107295 EZA53106.1 AAAB01008980 EAA14313.3 KA648027 AFP62656.1 APCN01001728 KZ288269 PBC30023.1 GGMS01013199 MBY82402.1 JRES01000841 KNC27815.1 UYSL01006581 VDL67534.1 GFDG01000452 JAV18347.1 GL441773 EFN64308.1 ATLV01021515 KE525319 KFB46637.1 GFXV01002749 MBW14554.1 UZAH01025668 VDO68344.1 GANO01003365 JAB56506.1 GCES01125253 JAQ61069.1 KZ355594 PIO60384.1 GCES01006432 JAR79891.1 GAMD01001056 JAB00535.1 UZAJ01000018 VDO24431.1 LBMM01015885 KMQ84631.1 GAMD01002902 JAA98688.1 GGFK01009372 MBW42693.1 GGFK01009359 MBW42680.1 GGFK01009386 MBW42707.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000283053
UP000075809
UP000007266
+ More
UP000007798 UP000007151 UP000000304 UP000000803 UP000008711 UP000001292 UP000192221 UP000076408 UP000053825 UP000001070 UP000002282 UP000078540 UP000092461 UP000008744 UP000005205 UP000007801 UP000268350 UP000075883 UP000008792 UP000076502 UP000001819 UP000078492 UP000075884 UP000053105 UP000009192 UP000075901 UP000075880 UP000007755 UP000078200 UP000092445 UP000092444 UP000078541 UP000075886 UP000092443 UP000092460 UP000091820 UP000037510 UP000075920 UP000075900 UP000008237 UP000053097 UP000007062 UP000075881 UP000075885 UP000095301 UP000075840 UP000075903 UP000076407 UP000075882 UP000075902 UP000242457 UP000005203 UP000037069 UP000095286 UP000038043 UP000271162 UP000095300 UP000000311 UP000030765 UP000095287 UP000192220 UP000050787 UP000036403 UP000265000 UP000265080 UP000264800 UP000095281
UP000007798 UP000007151 UP000000304 UP000000803 UP000008711 UP000001292 UP000192221 UP000076408 UP000053825 UP000001070 UP000002282 UP000078540 UP000092461 UP000008744 UP000005205 UP000007801 UP000268350 UP000075883 UP000008792 UP000076502 UP000001819 UP000078492 UP000075884 UP000053105 UP000009192 UP000075901 UP000075880 UP000007755 UP000078200 UP000092445 UP000092444 UP000078541 UP000075886 UP000092443 UP000092460 UP000091820 UP000037510 UP000075920 UP000075900 UP000008237 UP000053097 UP000007062 UP000075881 UP000075885 UP000095301 UP000075840 UP000075903 UP000076407 UP000075882 UP000075902 UP000242457 UP000005203 UP000037069 UP000095286 UP000038043 UP000271162 UP000095300 UP000000311 UP000030765 UP000095287 UP000192220 UP000050787 UP000036403 UP000265000 UP000265080 UP000264800 UP000095281
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
A0A2A4J158
A0A194R3G6
A0A194PNF7
A0A2H1VE26
A0A3S2N3P1
S4PX69
+ More
A0A151WU50 D6X0E4 B4MV91 A0A212EU15 B4Q490 A0A0J9R1G7 Q9VK62 B3N3C9 M9PD12 B4IEC0 A0A1W4UT61 A0A182YDJ4 A0A0L7RDP4 B4JQD7 A0A1B6FJ15 B4P2I6 A0A195AZ37 A0A1B0CKY0 B4HB27 A0A158NYD7 B3MNJ0 A0A3B0JIB4 A0A182M406 A0A310SP55 B4MDM3 A0A154PB38 Q29L74 A0A151J5D7 A0A182NC11 E9JD25 A0A0M8ZYI2 A0A0A1XQX0 B4KIN1 A0A182T0N9 A0A336N1L2 A0A182JBC5 W8BRX5 A0A034WQ74 F4WSY5 A0A1A9VMF4 A0A1B0AEN7 D3TMV7 A0A0T6BF60 A0A1B6ML95 A0A195F7H0 A0A182QRT6 A0A1A9Y3K0 A0A1B0BLA1 A0A1A9WM35 A0A0L7KRJ4 A0A182WGR3 A0A182RBJ4 E2BE87 A0A026WBW2 Q7Q1D5 T1PGV5 A0A182JUG2 A0A182PLM8 A0A1I8M2M9 A0A182HRN8 A0A182V3J9 A0A182X346 A0A182KTS3 A0A182UBR9 A0A2A3EE13 A0A088AN02 A0A2S2QZ46 A0A0L0C6A6 A0A1I8CKL1 A0A0N4XN42 A0A1L8EI08 A0A1I8PRC3 E2AQJ9 A0A084W8U3 A0A2H8TK96 A0A1I7ZKV4 A0A2I4AUU5 A0A3P7YAY7 U5ES80 A0A146QY91 A0A2G9TQX1 A0A147AN85 T1DK97 A0A183GXW8 A0A0J7K383 A0A3Q2PJM6 T1E7M5 A0A3P8S3P4 A0A3Q3BBT7 A0A3P8S1D6 A0A1I8BLD1 A0A2M4APJ6 A0A2M4API0 A0A2M4APM3
A0A151WU50 D6X0E4 B4MV91 A0A212EU15 B4Q490 A0A0J9R1G7 Q9VK62 B3N3C9 M9PD12 B4IEC0 A0A1W4UT61 A0A182YDJ4 A0A0L7RDP4 B4JQD7 A0A1B6FJ15 B4P2I6 A0A195AZ37 A0A1B0CKY0 B4HB27 A0A158NYD7 B3MNJ0 A0A3B0JIB4 A0A182M406 A0A310SP55 B4MDM3 A0A154PB38 Q29L74 A0A151J5D7 A0A182NC11 E9JD25 A0A0M8ZYI2 A0A0A1XQX0 B4KIN1 A0A182T0N9 A0A336N1L2 A0A182JBC5 W8BRX5 A0A034WQ74 F4WSY5 A0A1A9VMF4 A0A1B0AEN7 D3TMV7 A0A0T6BF60 A0A1B6ML95 A0A195F7H0 A0A182QRT6 A0A1A9Y3K0 A0A1B0BLA1 A0A1A9WM35 A0A0L7KRJ4 A0A182WGR3 A0A182RBJ4 E2BE87 A0A026WBW2 Q7Q1D5 T1PGV5 A0A182JUG2 A0A182PLM8 A0A1I8M2M9 A0A182HRN8 A0A182V3J9 A0A182X346 A0A182KTS3 A0A182UBR9 A0A2A3EE13 A0A088AN02 A0A2S2QZ46 A0A0L0C6A6 A0A1I8CKL1 A0A0N4XN42 A0A1L8EI08 A0A1I8PRC3 E2AQJ9 A0A084W8U3 A0A2H8TK96 A0A1I7ZKV4 A0A2I4AUU5 A0A3P7YAY7 U5ES80 A0A146QY91 A0A2G9TQX1 A0A147AN85 T1DK97 A0A183GXW8 A0A0J7K383 A0A3Q2PJM6 T1E7M5 A0A3P8S3P4 A0A3Q3BBT7 A0A3P8S1D6 A0A1I8BLD1 A0A2M4APJ6 A0A2M4API0 A0A2M4APM3
Ontologies
GO
PANTHER
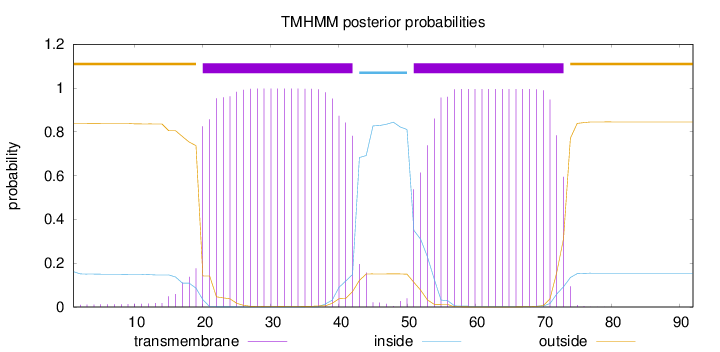
Topology
Length:
92
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
44.14545
Exp number, first 60 AAs:
31.76312
Total prob of N-in:
0.16112
POSSIBLE N-term signal
sequence
outside
1 - 19
TMhelix
20 - 42
inside
43 - 50
TMhelix
51 - 73
outside
74 - 92
Population Genetic Test Statistics
Pi
0.457725
Theta
4.166933
Tajima's D
-2.363887
CLR
1157.7744
CSRT
0.000999950002499875
Interpretation
Possibly Positive selection
