Pre Gene Modal
BGIBMGA010620
Annotation
syntaxin_5A_[Bombyx_mori]
Full name
Syntaxin-5
Alternative Name
Sed5 protein
dSed5
dSed5
Location in the cell
Mitochondrial Reliability : 1.395 Nuclear Reliability : 2.066
Sequence
CDS
ATGCTTCCTCGAAGGCGAAATATAGGCGTTACAGAAAGACTCTTGGAAACAGAAACGGATTCATACAATAAATTAGATAAGAAAGAAGAATTTGTTTTTGAACCAGTTATGGCTTCAAGAGATAGAACAAGTGAATTCATTTCAACTGTACGCAGTCTCCAAGGACGTTTCTTAAACAAACCAACCGTAAGAGATGATAGAAAAGCTGCTGTGTTGGAAACTTACTCCCAATTCATGAGTATGGCAAAAGTAATTAGTAAGAATATTACTAGTACATATACCAAATTGGAAAAGTTAGCCTTGTTGGCTAAGAGGAAATCATTATTTGATGACCGTCCCACAGAAATTCAAGAACTAACTTATATTATTAAAGGAGACTTAAATTCTTTAAATCAACAAATTGCCAGACTAGGTGAAATGCCAAGAGGTCGTCGCAGTATGCACAGTCATTCATCTAGTGTTGTACTCGCATTACAGTCACGCCTTGCTTCAATGAGTAATCAATTTAAACAGGTCTTAGAAGTTCGTTCAGAAAATTTGAAGCAACAGAACAGCAGGCGTGAACAATTTTCAAGGGTGACACCGGTTGTAAAAGAGGTACCATCTCTATTGCAACAAGATGAAGTAAGCATAGATTTGGGTGAAGCTACAAGTTTACAAGCACAGCAATTTGCATTCAGAGATGATACAGACTCATATGTACAACAGAGAGCTGAAACTATGCATAACATTGAAAGCACTATTGTGGAGTTGGGAGGCATATTTCAACAATTGGCTCATATGGTAAAAGAACAAGATGAAGCTATTGGTAGAATTGATGCAAACATTCATGAGGCAGAAATGAATGTTGAAGCTGGCCACAGAGAAATATTGAAATATTTCCAAAATGTAACTGGAAATAGGGCGTTAATGTTTAAAGTTTTTGGGGTTTTAATATTCTTTTTCATATTTTTTGTGGTGTTTATGGCTTAA
Protein
MLPRRRNIGVTERLLETETDSYNKLDKKEEFVFEPVMASRDRTSEFISTVRSLQGRFLNKPTVRDDRKAAVLETYSQFMSMAKVISKNITSTYTKLEKLALLAKRKSLFDDRPTEIQELTYIIKGDLNSLNQQIARLGEMPRGRRSMHSHSSSVVLALQSRLASMSNQFKQVLEVRSENLKQQNSRREQFSRVTPVVKEVPSLLQQDEVSIDLGEATSLQAQQFAFRDDTDSYVQQRAETMHNIESTIVELGGIFQQLAHMVKEQDEAIGRIDANIHEAEMNVEAGHREILKYFQNVTGNRALMFKVFGVLIFFFIFFVVFMA
Summary
Description
Mediates endoplasmic reticulum to Golgi transport.
Subunit
Homodimer.
Similarity
Belongs to the syntaxin family.
Belongs to the cullin family.
Belongs to the cullin family.
Keywords
Coiled coil
Complete proteome
ER-Golgi transport
Golgi apparatus
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Transport
Feature
chain Syntaxin-5
Uniprot
H9JM67
Q1HQ40
A0A3S2LPV2
A0A2H1VE62
A0A194PGY4
A0A2A4J0R1
+ More
A0A0N1IET6 A0A1E1WLZ8 A0A0L7LJC8 A0A212EU01 S4PGC9 A0A1J1IJJ4 A0A0K8TPE5 A0A2M3Z5E0 A0A2M3Z5E2 A0A2M4A977 A0A1W4X1W2 B4NXP5 A0A1W4VXZ9 W5JQT0 A0A2M4BNY1 A0A2M4BNJ5 A0A182F1I6 A0A0J9R2N6 A4V0S1 Q24509 B4I216 B4KI21 B3N660 A0A2J7PMX0 A0A2P8YVI3 B3MJN9 A0A3B0JGJ7 B0W6Z0 A0A0L0CC41 A0A067RN12 A0A182RVR5 A0A1I8MP04 A0A182VTK4 A0A1I8Q4F3 U5ERI5 A0A182P2U7 A0A1B0C025 A0A1A9Y5B6 A0A182NB86 B4GQN1 Q29NM4 A0A182SPP9 A0A1A9WM30 B4MU60 A0A182JII3 A0A1B6BXF3 A0A182QTU8 A0A1Y1K3U5 A0A1L8DVD0 A0A084VMP3 A0A182YM18 A0A182M109 A0A1B0A3Y4 A0A034W3F3 B4LTD5 A0A0K8VXJ5 A0A0A1WT30 B4Q700 A0A182UGM7 A0A0M4EGE1 A0A182KWL5 A0A182K7M8 A0A1A9UR06 A0A182VCS8 W8BWL3 A0A182G2K4 D3TPX4 V5IA44 A0A182IB78 A0A182X656 A0A1S4GZ62 B4JAB8 A0A0T6AYM1 J3JWV9 A0A336KM01 A0A0K8STS9 A0A146L5A7 A0A0C9RJL3 A0A182GUF8 A0A0L7RJC8 T1JGD8 E0VP35 A0A0V0G2X3 A0A069DSZ1 A0A224XT20 A0A2C9KBG1 T1I8W4 A0A0P4VL18 E2AXV7 U4UDG3 A0A026WM58 A0A154P0D0 M7BCC2 A0A0B7ATL4
A0A0N1IET6 A0A1E1WLZ8 A0A0L7LJC8 A0A212EU01 S4PGC9 A0A1J1IJJ4 A0A0K8TPE5 A0A2M3Z5E0 A0A2M3Z5E2 A0A2M4A977 A0A1W4X1W2 B4NXP5 A0A1W4VXZ9 W5JQT0 A0A2M4BNY1 A0A2M4BNJ5 A0A182F1I6 A0A0J9R2N6 A4V0S1 Q24509 B4I216 B4KI21 B3N660 A0A2J7PMX0 A0A2P8YVI3 B3MJN9 A0A3B0JGJ7 B0W6Z0 A0A0L0CC41 A0A067RN12 A0A182RVR5 A0A1I8MP04 A0A182VTK4 A0A1I8Q4F3 U5ERI5 A0A182P2U7 A0A1B0C025 A0A1A9Y5B6 A0A182NB86 B4GQN1 Q29NM4 A0A182SPP9 A0A1A9WM30 B4MU60 A0A182JII3 A0A1B6BXF3 A0A182QTU8 A0A1Y1K3U5 A0A1L8DVD0 A0A084VMP3 A0A182YM18 A0A182M109 A0A1B0A3Y4 A0A034W3F3 B4LTD5 A0A0K8VXJ5 A0A0A1WT30 B4Q700 A0A182UGM7 A0A0M4EGE1 A0A182KWL5 A0A182K7M8 A0A1A9UR06 A0A182VCS8 W8BWL3 A0A182G2K4 D3TPX4 V5IA44 A0A182IB78 A0A182X656 A0A1S4GZ62 B4JAB8 A0A0T6AYM1 J3JWV9 A0A336KM01 A0A0K8STS9 A0A146L5A7 A0A0C9RJL3 A0A182GUF8 A0A0L7RJC8 T1JGD8 E0VP35 A0A0V0G2X3 A0A069DSZ1 A0A224XT20 A0A2C9KBG1 T1I8W4 A0A0P4VL18 E2AXV7 U4UDG3 A0A026WM58 A0A154P0D0 M7BCC2 A0A0B7ATL4
Pubmed
19121390
26354079
26227816
22118469
23622113
26369729
+ More
17994087 17550304 20920257 23761445 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10471707 12537569 7929581 29403074 18057021 26108605 24845553 25315136 15632085 28004739 24438588 25244985 25348373 25830018 20966253 24495485 26483478 20353571 12364791 22516182 26823975 20566863 26334808 15562597 27129103 20798317 23537049 24508170 30249741 23624526
17994087 17550304 20920257 23761445 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10471707 12537569 7929581 29403074 18057021 26108605 24845553 25315136 15632085 28004739 24438588 25244985 25348373 25830018 20966253 24495485 26483478 20353571 12364791 22516182 26823975 20566863 26334808 15562597 27129103 20798317 23537049 24508170 30249741 23624526
EMBL
BABH01033436
BABH01033437
DQ443212
ABF51301.1
RSAL01000762
RVE41167.1
+ More
ODYU01002047 SOQ39066.1 KQ459603 KPI92656.1 NWSH01004441 PCG65114.1 KQ460456 KPJ14661.1 GDQN01003001 JAT88053.1 JTDY01000955 KOB75296.1 AGBW02012509 OWR44970.1 GAIX01003712 JAA88848.1 CVRI01000054 CRL00415.1 GDAI01001580 JAI16023.1 GGFM01002981 MBW23732.1 GGFM01002982 MBW23733.1 GGFK01004032 MBW37353.1 CM000157 EDW89670.1 KRJ98651.1 ADMH02000444 ETN66456.1 GGFJ01005510 MBW54651.1 GGFJ01005509 MBW54650.1 CM002910 KMY90496.1 KMY90497.1 AE014134 AAN10935.1 AY069832 X78219 AAF53520.2 AAL39977.1 CAA55064.1 CH480820 EDW54573.1 CH933807 EDW11303.1 CH954177 EDV58098.1 NEVH01023960 PNF17684.1 PYGN01000337 PSN48254.1 CH902620 EDV31378.1 KPU73342.1 KPU73343.1 OUUW01000006 SPP81477.1 DS231851 EDS37264.1 JRES01000714 KNC29009.1 KK852572 KDR21104.1 GANO01003727 JAB56144.1 JXJN01023462 CH479187 EDW39903.1 CH379059 EAL34194.1 CH963852 EDW75649.2 GEDC01031663 GEDC01001151 JAS05635.1 JAS36147.1 AXCN02000633 GEZM01093798 JAV56122.1 GFDF01003686 JAV10398.1 ATLV01014616 KE524975 KFB39237.1 AXCM01000488 GAKP01010659 JAC48293.1 CH940649 EDW63905.1 GDHF01009069 JAI43245.1 GBXI01012632 JAD01660.1 CM000361 EDX05208.1 CP012523 ALC39755.1 GAMC01005262 JAC01294.1 JXUM01139644 JXUM01139645 JXUM01139646 JXUM01139647 JXUM01139648 KQ568932 KXJ68833.1 CCAG010009633 EZ423476 ADD19752.1 GALX01001830 JAB66636.1 APCN01000644 AAAB01008944 CH916368 EDW03789.1 LJIG01022493 KRT80287.1 BT127727 AEE62689.1 UFQS01000439 UFQT01000439 SSX03953.1 SSX24318.1 GBRD01009544 JAG56280.1 GDHC01015026 GDHC01007955 JAQ03603.1 JAQ10674.1 GBYB01007061 GBYB01011406 JAG76828.1 JAG81173.1 JXUM01089073 JXUM01089074 JXUM01089075 JXUM01089076 JXUM01089077 KQ563744 KXJ73397.1 KQ414579 KOC71077.1 JH432201 DS235354 EEB15141.1 GECL01003626 JAP02498.1 GBGD01002107 JAC86782.1 GFTR01005163 JAW11263.1 ACPB03001979 ACPB03001980 GDKW01003549 JAI53046.1 GL443736 EFN61721.1 KB632308 ERL92014.1 KK107168 QOIP01000001 EZA56189.1 RLU27429.1 KQ434782 KZC04600.1 KB586437 EMP25857.1 HACG01037057 HACG01037060 CEK83922.1 CEK83925.1
ODYU01002047 SOQ39066.1 KQ459603 KPI92656.1 NWSH01004441 PCG65114.1 KQ460456 KPJ14661.1 GDQN01003001 JAT88053.1 JTDY01000955 KOB75296.1 AGBW02012509 OWR44970.1 GAIX01003712 JAA88848.1 CVRI01000054 CRL00415.1 GDAI01001580 JAI16023.1 GGFM01002981 MBW23732.1 GGFM01002982 MBW23733.1 GGFK01004032 MBW37353.1 CM000157 EDW89670.1 KRJ98651.1 ADMH02000444 ETN66456.1 GGFJ01005510 MBW54651.1 GGFJ01005509 MBW54650.1 CM002910 KMY90496.1 KMY90497.1 AE014134 AAN10935.1 AY069832 X78219 AAF53520.2 AAL39977.1 CAA55064.1 CH480820 EDW54573.1 CH933807 EDW11303.1 CH954177 EDV58098.1 NEVH01023960 PNF17684.1 PYGN01000337 PSN48254.1 CH902620 EDV31378.1 KPU73342.1 KPU73343.1 OUUW01000006 SPP81477.1 DS231851 EDS37264.1 JRES01000714 KNC29009.1 KK852572 KDR21104.1 GANO01003727 JAB56144.1 JXJN01023462 CH479187 EDW39903.1 CH379059 EAL34194.1 CH963852 EDW75649.2 GEDC01031663 GEDC01001151 JAS05635.1 JAS36147.1 AXCN02000633 GEZM01093798 JAV56122.1 GFDF01003686 JAV10398.1 ATLV01014616 KE524975 KFB39237.1 AXCM01000488 GAKP01010659 JAC48293.1 CH940649 EDW63905.1 GDHF01009069 JAI43245.1 GBXI01012632 JAD01660.1 CM000361 EDX05208.1 CP012523 ALC39755.1 GAMC01005262 JAC01294.1 JXUM01139644 JXUM01139645 JXUM01139646 JXUM01139647 JXUM01139648 KQ568932 KXJ68833.1 CCAG010009633 EZ423476 ADD19752.1 GALX01001830 JAB66636.1 APCN01000644 AAAB01008944 CH916368 EDW03789.1 LJIG01022493 KRT80287.1 BT127727 AEE62689.1 UFQS01000439 UFQT01000439 SSX03953.1 SSX24318.1 GBRD01009544 JAG56280.1 GDHC01015026 GDHC01007955 JAQ03603.1 JAQ10674.1 GBYB01007061 GBYB01011406 JAG76828.1 JAG81173.1 JXUM01089073 JXUM01089074 JXUM01089075 JXUM01089076 JXUM01089077 KQ563744 KXJ73397.1 KQ414579 KOC71077.1 JH432201 DS235354 EEB15141.1 GECL01003626 JAP02498.1 GBGD01002107 JAC86782.1 GFTR01005163 JAW11263.1 ACPB03001979 ACPB03001980 GDKW01003549 JAI53046.1 GL443736 EFN61721.1 KB632308 ERL92014.1 KK107168 QOIP01000001 EZA56189.1 RLU27429.1 KQ434782 KZC04600.1 KB586437 EMP25857.1 HACG01037057 HACG01037060 CEK83922.1 CEK83925.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000218220
UP000053240
UP000037510
+ More
UP000007151 UP000183832 UP000192223 UP000002282 UP000192221 UP000000673 UP000069272 UP000000803 UP000001292 UP000009192 UP000008711 UP000235965 UP000245037 UP000007801 UP000268350 UP000002320 UP000037069 UP000027135 UP000075900 UP000095301 UP000075920 UP000095300 UP000075885 UP000092460 UP000092443 UP000075884 UP000008744 UP000001819 UP000075901 UP000091820 UP000007798 UP000075880 UP000075886 UP000030765 UP000076408 UP000075883 UP000092445 UP000008792 UP000000304 UP000075902 UP000092553 UP000075882 UP000075881 UP000078200 UP000075903 UP000069940 UP000249989 UP000092444 UP000075840 UP000076407 UP000001070 UP000053825 UP000009046 UP000076420 UP000015103 UP000000311 UP000030742 UP000053097 UP000279307 UP000076502 UP000031443
UP000007151 UP000183832 UP000192223 UP000002282 UP000192221 UP000000673 UP000069272 UP000000803 UP000001292 UP000009192 UP000008711 UP000235965 UP000245037 UP000007801 UP000268350 UP000002320 UP000037069 UP000027135 UP000075900 UP000095301 UP000075920 UP000095300 UP000075885 UP000092460 UP000092443 UP000075884 UP000008744 UP000001819 UP000075901 UP000091820 UP000007798 UP000075880 UP000075886 UP000030765 UP000076408 UP000075883 UP000092445 UP000008792 UP000000304 UP000075902 UP000092553 UP000075882 UP000075881 UP000078200 UP000075903 UP000069940 UP000249989 UP000092444 UP000075840 UP000076407 UP000001070 UP000053825 UP000009046 UP000076420 UP000015103 UP000000311 UP000030742 UP000053097 UP000279307 UP000076502 UP000031443
Pfam
Interpro
IPR010989
SNARE
+ More
IPR006011 Syntaxin_N
IPR000727 T_SNARE_dom
IPR021538 Syntaxin-5_N
IPR039726 Prp40-like
IPR036020 WW_dom_sf
IPR001202 WW_dom
IPR002713 FF_domain
IPR036517 FF_domain_sf
IPR006012 Syntaxin/epimorphin_CS
IPR016159 Cullin_repeat-like_dom_sf
IPR016158 Cullin_homology
IPR036317 Cullin_homology_sf
IPR019559 Cullin_neddylation_domain
IPR036390 WH_DNA-bd_sf
IPR001373 Cullin_N
IPR036388 WH-like_DNA-bd_sf
IPR006011 Syntaxin_N
IPR000727 T_SNARE_dom
IPR021538 Syntaxin-5_N
IPR039726 Prp40-like
IPR036020 WW_dom_sf
IPR001202 WW_dom
IPR002713 FF_domain
IPR036517 FF_domain_sf
IPR006012 Syntaxin/epimorphin_CS
IPR016159 Cullin_repeat-like_dom_sf
IPR016158 Cullin_homology
IPR036317 Cullin_homology_sf
IPR019559 Cullin_neddylation_domain
IPR036390 WH_DNA-bd_sf
IPR001373 Cullin_N
IPR036388 WH-like_DNA-bd_sf
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9JM67
Q1HQ40
A0A3S2LPV2
A0A2H1VE62
A0A194PGY4
A0A2A4J0R1
+ More
A0A0N1IET6 A0A1E1WLZ8 A0A0L7LJC8 A0A212EU01 S4PGC9 A0A1J1IJJ4 A0A0K8TPE5 A0A2M3Z5E0 A0A2M3Z5E2 A0A2M4A977 A0A1W4X1W2 B4NXP5 A0A1W4VXZ9 W5JQT0 A0A2M4BNY1 A0A2M4BNJ5 A0A182F1I6 A0A0J9R2N6 A4V0S1 Q24509 B4I216 B4KI21 B3N660 A0A2J7PMX0 A0A2P8YVI3 B3MJN9 A0A3B0JGJ7 B0W6Z0 A0A0L0CC41 A0A067RN12 A0A182RVR5 A0A1I8MP04 A0A182VTK4 A0A1I8Q4F3 U5ERI5 A0A182P2U7 A0A1B0C025 A0A1A9Y5B6 A0A182NB86 B4GQN1 Q29NM4 A0A182SPP9 A0A1A9WM30 B4MU60 A0A182JII3 A0A1B6BXF3 A0A182QTU8 A0A1Y1K3U5 A0A1L8DVD0 A0A084VMP3 A0A182YM18 A0A182M109 A0A1B0A3Y4 A0A034W3F3 B4LTD5 A0A0K8VXJ5 A0A0A1WT30 B4Q700 A0A182UGM7 A0A0M4EGE1 A0A182KWL5 A0A182K7M8 A0A1A9UR06 A0A182VCS8 W8BWL3 A0A182G2K4 D3TPX4 V5IA44 A0A182IB78 A0A182X656 A0A1S4GZ62 B4JAB8 A0A0T6AYM1 J3JWV9 A0A336KM01 A0A0K8STS9 A0A146L5A7 A0A0C9RJL3 A0A182GUF8 A0A0L7RJC8 T1JGD8 E0VP35 A0A0V0G2X3 A0A069DSZ1 A0A224XT20 A0A2C9KBG1 T1I8W4 A0A0P4VL18 E2AXV7 U4UDG3 A0A026WM58 A0A154P0D0 M7BCC2 A0A0B7ATL4
A0A0N1IET6 A0A1E1WLZ8 A0A0L7LJC8 A0A212EU01 S4PGC9 A0A1J1IJJ4 A0A0K8TPE5 A0A2M3Z5E0 A0A2M3Z5E2 A0A2M4A977 A0A1W4X1W2 B4NXP5 A0A1W4VXZ9 W5JQT0 A0A2M4BNY1 A0A2M4BNJ5 A0A182F1I6 A0A0J9R2N6 A4V0S1 Q24509 B4I216 B4KI21 B3N660 A0A2J7PMX0 A0A2P8YVI3 B3MJN9 A0A3B0JGJ7 B0W6Z0 A0A0L0CC41 A0A067RN12 A0A182RVR5 A0A1I8MP04 A0A182VTK4 A0A1I8Q4F3 U5ERI5 A0A182P2U7 A0A1B0C025 A0A1A9Y5B6 A0A182NB86 B4GQN1 Q29NM4 A0A182SPP9 A0A1A9WM30 B4MU60 A0A182JII3 A0A1B6BXF3 A0A182QTU8 A0A1Y1K3U5 A0A1L8DVD0 A0A084VMP3 A0A182YM18 A0A182M109 A0A1B0A3Y4 A0A034W3F3 B4LTD5 A0A0K8VXJ5 A0A0A1WT30 B4Q700 A0A182UGM7 A0A0M4EGE1 A0A182KWL5 A0A182K7M8 A0A1A9UR06 A0A182VCS8 W8BWL3 A0A182G2K4 D3TPX4 V5IA44 A0A182IB78 A0A182X656 A0A1S4GZ62 B4JAB8 A0A0T6AYM1 J3JWV9 A0A336KM01 A0A0K8STS9 A0A146L5A7 A0A0C9RJL3 A0A182GUF8 A0A0L7RJC8 T1JGD8 E0VP35 A0A0V0G2X3 A0A069DSZ1 A0A224XT20 A0A2C9KBG1 T1I8W4 A0A0P4VL18 E2AXV7 U4UDG3 A0A026WM58 A0A154P0D0 M7BCC2 A0A0B7ATL4
PDB
1GL2
E-value=0.021736,
Score=88
Ontologies
GO
GO:0016192
GO:0016021
GO:0045292
GO:0016020
GO:0006886
GO:0005623
GO:0005484
GO:0000902
GO:0007283
GO:0060305
GO:0035167
GO:0042803
GO:0000281
GO:0007112
GO:0008283
GO:0007030
GO:0009306
GO:0006887
GO:0005801
GO:0006878
GO:0045930
GO:0016081
GO:0012505
GO:0007269
GO:0000149
GO:0000139
GO:0005794
GO:0005886
GO:0055070
GO:0098793
GO:0005783
GO:0031201
GO:0006511
GO:0031625
GO:0005515
GO:0006487
GO:0006488
GO:0007156
GO:0008237
GO:0043401
GO:0003707
GO:0051537
PANTHER
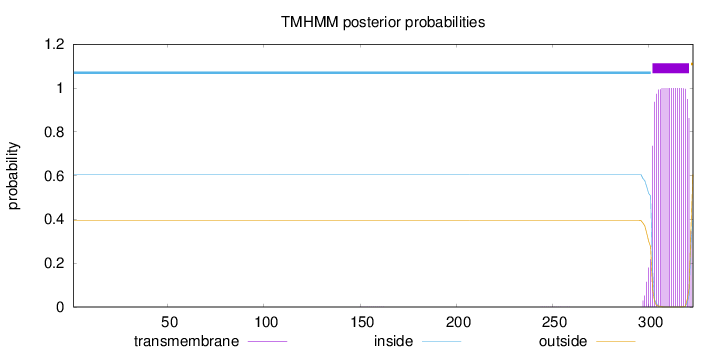
Topology
Subcellular location
Length:
323
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.38709
Exp number, first 60 AAs:
0
Total prob of N-in:
0.60513
inside
1 - 301
TMhelix
302 - 321
outside
322 - 323
Population Genetic Test Statistics
Pi
0.420184
Theta
1.925899
Tajima's D
-1.492888
CLR
1089.441074
CSRT
0.0495975201239938
Interpretation
Possibly Positive selection
