Gene
KWMTBOMO07328 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010668
Annotation
PREDICTED:_barrier-to-autointegration_factor_B_[Plutella_xylostella]
Full name
Barrier-to-autointegration factor B
Location in the cell
Cytoplasmic Reliability : 1.319 Extracellular Reliability : 1.041 Nuclear Reliability : 1.092
Sequence
CDS
ATGTCGAGTACATCACAAAAACATAGAAACTTTGTTGCTGAGCCAATGGGTGAGAAACCAGTAACAGAATTGGCGGGCGTGGGTGAAGTTTTAGGAAAACGACTTGAAACCGGGGGCTTCGATAAGGCCTATGTCGTCTTGGGTCAATTTTTGGTACTAAAGAAAGATAAAGAGTTATTTCAAGAATGGATGAAGGACGCCTGCAGTGCAAACTCTAAACAGTCAGCAGATTGTTATCAGTGTTTGAAGGATTGGTGTGATGAGTTTTTGTAA
Protein
MSSTSQKHRNFVAEPMGEKPVTELAGVGEVLGKRLETGGFDKAYVVLGQFLVLKKDKELFQEWMKDACSANSKQSADCYQCLKDWCDEFL
Summary
Description
Plays fundamental roles in nuclear assembly, chromatin organization, gene expression and gonad development. May potently compress chromatin structure and be involved in membrane recruitment and chromatin decondensation during nuclear assembly. Contains 2 non-specific dsDNA-binding sites which may promote DNA cross-bridging (By similarity).
Subunit
Homodimer. Binds non-specifically to double-stranded DNA, and is found as hexamer or dodecamer upon DNA binding. Binds to LEM domain-containing nuclear proteins (By similarity). Interacts with nemp1a and nemp1b.
Similarity
Belongs to the BAF family.
Keywords
Chromosome
DNA-binding
Nucleus
Phosphoprotein
Feature
chain Barrier-to-autointegration factor B
Uniprot
H9JMB5
A0A2H1WQB1
A0A0N1PJH5
A0A194PH77
S4PMT5
A0A2W1BK97
+ More
A0A2A4K0Q7 A0A3S2NFE2 A0A151I141 A0A158NG20 A0A195E805 A0A151XB73 F4X6K3 E2A9L1 A0A023FA51 E2BN03 A0A026WSU2 A0A232FLN1 K7IVY9 A0A0J7KL26 A0A0V0G9R1 A0A0M8ZXE7 A0A2P8YDF6 A0A069DNU4 A0A0L7LIL8 A0A2R7WXE5 D2A041 A0A154P269 A0A088AKM9 A0A151ILJ9 A0A310SJK4 A0A182QRX6 T1DPJ3 A0A2M4CTB1 A0A2M3ZKD5 A0A2M4C4X1 A0A182F3P2 A0A2J7PMW7 A0A182YM16 A0A2C9GS31 A0A182P2V0 A0A182MBC2 A0A182V206 A0A182K306 A0A182TE78 A0A182VTK1 A0A084VMP1 A0A182X659 Q7Q881 A0A182R441 A0A2M4AZ37 A0A195F1Z9 G1K0C4 A0A2A3EDU8 A0A1W4XWA5 A0A182NB89 E9J5Y3 T1DFU6 A0A0P4VRZ0 A0A1L8DX15 A0A1B0CDL9 A0A1Q3F4W9 B0W6Y7 N6UAN5 Q1HRI3 A0A023ECD3 U5ELZ8 A0A224XZD6 A0A2R5LHF0 A0A067RDK3 A0A023G0W2 A0A224YVQ0 A0A131YEX3 A0A131XN04 A0A023FDQ5 E0W3Y0 A0A0K8R5J2 A0A1E1WWF0 A0A1J1IJZ8 A0A182JHN6 C1BUT6 C1C252 A0A0K8TT20 C1BQK5 A0A2I4BVX9 A0A182KWK4 A0A3P8VU86 A0A3Q3E8F5 A0A3B4ZDG9 A0A3B3HU08 A0A3P9MLN3 A0A3Q3LEY2 A0A087T752 A0A3B4YLL8 A0A3B4UU04 A0A1L8GCL6 Q66KV4 A0A0E9X1H6 A0A1S3DEN4 A0A3Q3W3D2
A0A2A4K0Q7 A0A3S2NFE2 A0A151I141 A0A158NG20 A0A195E805 A0A151XB73 F4X6K3 E2A9L1 A0A023FA51 E2BN03 A0A026WSU2 A0A232FLN1 K7IVY9 A0A0J7KL26 A0A0V0G9R1 A0A0M8ZXE7 A0A2P8YDF6 A0A069DNU4 A0A0L7LIL8 A0A2R7WXE5 D2A041 A0A154P269 A0A088AKM9 A0A151ILJ9 A0A310SJK4 A0A182QRX6 T1DPJ3 A0A2M4CTB1 A0A2M3ZKD5 A0A2M4C4X1 A0A182F3P2 A0A2J7PMW7 A0A182YM16 A0A2C9GS31 A0A182P2V0 A0A182MBC2 A0A182V206 A0A182K306 A0A182TE78 A0A182VTK1 A0A084VMP1 A0A182X659 Q7Q881 A0A182R441 A0A2M4AZ37 A0A195F1Z9 G1K0C4 A0A2A3EDU8 A0A1W4XWA5 A0A182NB89 E9J5Y3 T1DFU6 A0A0P4VRZ0 A0A1L8DX15 A0A1B0CDL9 A0A1Q3F4W9 B0W6Y7 N6UAN5 Q1HRI3 A0A023ECD3 U5ELZ8 A0A224XZD6 A0A2R5LHF0 A0A067RDK3 A0A023G0W2 A0A224YVQ0 A0A131YEX3 A0A131XN04 A0A023FDQ5 E0W3Y0 A0A0K8R5J2 A0A1E1WWF0 A0A1J1IJZ8 A0A182JHN6 C1BUT6 C1C252 A0A0K8TT20 C1BQK5 A0A2I4BVX9 A0A182KWK4 A0A3P8VU86 A0A3Q3E8F5 A0A3B4ZDG9 A0A3B3HU08 A0A3P9MLN3 A0A3Q3LEY2 A0A087T752 A0A3B4YLL8 A0A3B4UU04 A0A1L8GCL6 Q66KV4 A0A0E9X1H6 A0A1S3DEN4 A0A3Q3W3D2
Pubmed
19121390
26354079
23622113
28756777
21347285
21719571
+ More
20798317 25474469 24508170 30249741 28648823 20075255 29403074 26334808 26227816 18362917 19820115 25244985 24438588 12364791 14747013 17210077 21736942 21282665 24330624 27129103 23537049 17204158 17510324 24945155 26483478 24845553 28797301 26830274 28049606 20566863 28503490 26369729 20966253 24487278 17554307 27762356 19167377 25613341
20798317 25474469 24508170 30249741 28648823 20075255 29403074 26334808 26227816 18362917 19820115 25244985 24438588 12364791 14747013 17210077 21736942 21282665 24330624 27129103 23537049 17204158 17510324 24945155 26483478 24845553 28797301 26830274 28049606 20566863 28503490 26369729 20966253 24487278 17554307 27762356 19167377 25613341
EMBL
BABH01033435
BABH01033436
ODYU01009901
SOQ54624.1
KQ460456
KPJ14662.1
+ More
KQ459603 KPI92657.1 GAIX01000006 JAA92554.1 KZ150090 PZC73697.1 NWSH01000276 PCG77827.1 RSAL01000139 RVE46117.1 KQ976580 KYM79899.1 ADTU01014889 KQ979568 KYN20957.1 KQ982335 KYQ57606.1 GL888818 EGI57963.1 GL437917 EFN69901.1 GBBI01000909 JAC17803.1 GL449382 EFN82907.1 KK107109 QOIP01000008 EZA59100.1 RLU19458.1 NNAY01000040 OXU31644.1 LBMM01006104 KMQ90936.1 GECL01002013 JAP04111.1 KQ435811 KOX72877.1 PYGN01000689 PSN42204.1 GBGD01003627 JAC85262.1 JTDY01000955 KOB75292.1 KK855873 PTY24208.1 KQ971338 EFA02465.1 KQ434787 KZC05308.1 KQ977104 KYN05765.1 KQ760551 OAD59838.1 OAD59839.1 AXCN02000633 GAMD01003299 JAA98291.1 GGFL01004389 MBW68567.1 GGFM01008169 MBW28920.1 GGFJ01011168 MBW60309.1 NEVH01023960 PNF17681.1 APCN01000645 AXCM01002111 ATLV01014616 KE524975 KFB39235.1 AAAB01008944 EAA10294.1 GGFK01012661 MBW45982.1 KQ981864 KYN34192.1 JO495005 AEL79235.1 KZ288285 PBC29462.1 GL768221 EFZ11768.1 GALA01000432 JAA94420.1 GDKW01001927 JAI54668.1 GFDF01003093 JAV10991.1 AJWK01007998 GFDL01012434 JAV22611.1 DS231851 EDS37261.1 APGK01032040 KB740834 KB632166 ENN78800.1 ERL89332.1 DQ440111 CH477527 ABF18144.1 EAT39517.1 JXUM01086049 GAPW01006624 GAPW01006620 KQ563540 JAC06978.1 KXJ73694.1 GANO01001212 JAB58659.1 GFTR01001008 JAW15418.1 GGLE01004621 MBY08747.1 KK852572 KDR21103.1 GBBL01000815 JAC26505.1 GFPF01007237 MAA18383.1 GEDV01011525 JAP77032.1 GEFH01000152 JAP68429.1 GBBK01004451 JAC20031.1 DS235883 EEB20336.1 GADI01007371 JAA66437.1 GFAC01007833 JAT91355.1 CVRI01000054 CRK99868.1 BT078365 ACO12789.1 BT080931 ACO15355.1 GDAI01000310 JAI17293.1 BT076884 BT076967 ACO11308.1 KK113747 KFM60941.1 CM004473 OCT81677.1 BC078546 GBXM01013019 JAH95558.1
KQ459603 KPI92657.1 GAIX01000006 JAA92554.1 KZ150090 PZC73697.1 NWSH01000276 PCG77827.1 RSAL01000139 RVE46117.1 KQ976580 KYM79899.1 ADTU01014889 KQ979568 KYN20957.1 KQ982335 KYQ57606.1 GL888818 EGI57963.1 GL437917 EFN69901.1 GBBI01000909 JAC17803.1 GL449382 EFN82907.1 KK107109 QOIP01000008 EZA59100.1 RLU19458.1 NNAY01000040 OXU31644.1 LBMM01006104 KMQ90936.1 GECL01002013 JAP04111.1 KQ435811 KOX72877.1 PYGN01000689 PSN42204.1 GBGD01003627 JAC85262.1 JTDY01000955 KOB75292.1 KK855873 PTY24208.1 KQ971338 EFA02465.1 KQ434787 KZC05308.1 KQ977104 KYN05765.1 KQ760551 OAD59838.1 OAD59839.1 AXCN02000633 GAMD01003299 JAA98291.1 GGFL01004389 MBW68567.1 GGFM01008169 MBW28920.1 GGFJ01011168 MBW60309.1 NEVH01023960 PNF17681.1 APCN01000645 AXCM01002111 ATLV01014616 KE524975 KFB39235.1 AAAB01008944 EAA10294.1 GGFK01012661 MBW45982.1 KQ981864 KYN34192.1 JO495005 AEL79235.1 KZ288285 PBC29462.1 GL768221 EFZ11768.1 GALA01000432 JAA94420.1 GDKW01001927 JAI54668.1 GFDF01003093 JAV10991.1 AJWK01007998 GFDL01012434 JAV22611.1 DS231851 EDS37261.1 APGK01032040 KB740834 KB632166 ENN78800.1 ERL89332.1 DQ440111 CH477527 ABF18144.1 EAT39517.1 JXUM01086049 GAPW01006624 GAPW01006620 KQ563540 JAC06978.1 KXJ73694.1 GANO01001212 JAB58659.1 GFTR01001008 JAW15418.1 GGLE01004621 MBY08747.1 KK852572 KDR21103.1 GBBL01000815 JAC26505.1 GFPF01007237 MAA18383.1 GEDV01011525 JAP77032.1 GEFH01000152 JAP68429.1 GBBK01004451 JAC20031.1 DS235883 EEB20336.1 GADI01007371 JAA66437.1 GFAC01007833 JAT91355.1 CVRI01000054 CRK99868.1 BT078365 ACO12789.1 BT080931 ACO15355.1 GDAI01000310 JAI17293.1 BT076884 BT076967 ACO11308.1 KK113747 KFM60941.1 CM004473 OCT81677.1 BC078546 GBXM01013019 JAH95558.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000283053
UP000078540
+ More
UP000005205 UP000078492 UP000075809 UP000007755 UP000000311 UP000008237 UP000053097 UP000279307 UP000215335 UP000002358 UP000036403 UP000053105 UP000245037 UP000037510 UP000007266 UP000076502 UP000005203 UP000078542 UP000075886 UP000069272 UP000235965 UP000076408 UP000075840 UP000075885 UP000075883 UP000075903 UP000075881 UP000075902 UP000075920 UP000030765 UP000076407 UP000007062 UP000075900 UP000078541 UP000242457 UP000192223 UP000075884 UP000092461 UP000002320 UP000019118 UP000030742 UP000008820 UP000069940 UP000249989 UP000027135 UP000009046 UP000183832 UP000075880 UP000192220 UP000075882 UP000265120 UP000261660 UP000261400 UP000001038 UP000265180 UP000265200 UP000261640 UP000054359 UP000261360 UP000261420 UP000186698 UP000079169 UP000261620
UP000005205 UP000078492 UP000075809 UP000007755 UP000000311 UP000008237 UP000053097 UP000279307 UP000215335 UP000002358 UP000036403 UP000053105 UP000245037 UP000037510 UP000007266 UP000076502 UP000005203 UP000078542 UP000075886 UP000069272 UP000235965 UP000076408 UP000075840 UP000075885 UP000075883 UP000075903 UP000075881 UP000075902 UP000075920 UP000030765 UP000076407 UP000007062 UP000075900 UP000078541 UP000242457 UP000192223 UP000075884 UP000092461 UP000002320 UP000019118 UP000030742 UP000008820 UP000069940 UP000249989 UP000027135 UP000009046 UP000183832 UP000075880 UP000192220 UP000075882 UP000265120 UP000261660 UP000261400 UP000001038 UP000265180 UP000265200 UP000261640 UP000054359 UP000261360 UP000261420 UP000186698 UP000079169 UP000261620
Pfam
PF02961 BAF
Interpro
Gene 3D
ProteinModelPortal
H9JMB5
A0A2H1WQB1
A0A0N1PJH5
A0A194PH77
S4PMT5
A0A2W1BK97
+ More
A0A2A4K0Q7 A0A3S2NFE2 A0A151I141 A0A158NG20 A0A195E805 A0A151XB73 F4X6K3 E2A9L1 A0A023FA51 E2BN03 A0A026WSU2 A0A232FLN1 K7IVY9 A0A0J7KL26 A0A0V0G9R1 A0A0M8ZXE7 A0A2P8YDF6 A0A069DNU4 A0A0L7LIL8 A0A2R7WXE5 D2A041 A0A154P269 A0A088AKM9 A0A151ILJ9 A0A310SJK4 A0A182QRX6 T1DPJ3 A0A2M4CTB1 A0A2M3ZKD5 A0A2M4C4X1 A0A182F3P2 A0A2J7PMW7 A0A182YM16 A0A2C9GS31 A0A182P2V0 A0A182MBC2 A0A182V206 A0A182K306 A0A182TE78 A0A182VTK1 A0A084VMP1 A0A182X659 Q7Q881 A0A182R441 A0A2M4AZ37 A0A195F1Z9 G1K0C4 A0A2A3EDU8 A0A1W4XWA5 A0A182NB89 E9J5Y3 T1DFU6 A0A0P4VRZ0 A0A1L8DX15 A0A1B0CDL9 A0A1Q3F4W9 B0W6Y7 N6UAN5 Q1HRI3 A0A023ECD3 U5ELZ8 A0A224XZD6 A0A2R5LHF0 A0A067RDK3 A0A023G0W2 A0A224YVQ0 A0A131YEX3 A0A131XN04 A0A023FDQ5 E0W3Y0 A0A0K8R5J2 A0A1E1WWF0 A0A1J1IJZ8 A0A182JHN6 C1BUT6 C1C252 A0A0K8TT20 C1BQK5 A0A2I4BVX9 A0A182KWK4 A0A3P8VU86 A0A3Q3E8F5 A0A3B4ZDG9 A0A3B3HU08 A0A3P9MLN3 A0A3Q3LEY2 A0A087T752 A0A3B4YLL8 A0A3B4UU04 A0A1L8GCL6 Q66KV4 A0A0E9X1H6 A0A1S3DEN4 A0A3Q3W3D2
A0A2A4K0Q7 A0A3S2NFE2 A0A151I141 A0A158NG20 A0A195E805 A0A151XB73 F4X6K3 E2A9L1 A0A023FA51 E2BN03 A0A026WSU2 A0A232FLN1 K7IVY9 A0A0J7KL26 A0A0V0G9R1 A0A0M8ZXE7 A0A2P8YDF6 A0A069DNU4 A0A0L7LIL8 A0A2R7WXE5 D2A041 A0A154P269 A0A088AKM9 A0A151ILJ9 A0A310SJK4 A0A182QRX6 T1DPJ3 A0A2M4CTB1 A0A2M3ZKD5 A0A2M4C4X1 A0A182F3P2 A0A2J7PMW7 A0A182YM16 A0A2C9GS31 A0A182P2V0 A0A182MBC2 A0A182V206 A0A182K306 A0A182TE78 A0A182VTK1 A0A084VMP1 A0A182X659 Q7Q881 A0A182R441 A0A2M4AZ37 A0A195F1Z9 G1K0C4 A0A2A3EDU8 A0A1W4XWA5 A0A182NB89 E9J5Y3 T1DFU6 A0A0P4VRZ0 A0A1L8DX15 A0A1B0CDL9 A0A1Q3F4W9 B0W6Y7 N6UAN5 Q1HRI3 A0A023ECD3 U5ELZ8 A0A224XZD6 A0A2R5LHF0 A0A067RDK3 A0A023G0W2 A0A224YVQ0 A0A131YEX3 A0A131XN04 A0A023FDQ5 E0W3Y0 A0A0K8R5J2 A0A1E1WWF0 A0A1J1IJZ8 A0A182JHN6 C1BUT6 C1C252 A0A0K8TT20 C1BQK5 A0A2I4BVX9 A0A182KWK4 A0A3P8VU86 A0A3Q3E8F5 A0A3B4ZDG9 A0A3B3HU08 A0A3P9MLN3 A0A3Q3LEY2 A0A087T752 A0A3B4YLL8 A0A3B4UU04 A0A1L8GCL6 Q66KV4 A0A0E9X1H6 A0A1S3DEN4 A0A3Q3W3D2
PDB
2ODG
E-value=1.14786e-34,
Score=360
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Nucleus
Colocalizes with nemp1a and nemp1b at the nuclear envelope. With evidence from 2 publications.
Nucleus envelope Colocalizes with nemp1a and nemp1b at the nuclear envelope. With evidence from 2 publications.
Chromosome Colocalizes with nemp1a and nemp1b at the nuclear envelope. With evidence from 2 publications.
Nucleus envelope Colocalizes with nemp1a and nemp1b at the nuclear envelope. With evidence from 2 publications.
Chromosome Colocalizes with nemp1a and nemp1b at the nuclear envelope. With evidence from 2 publications.
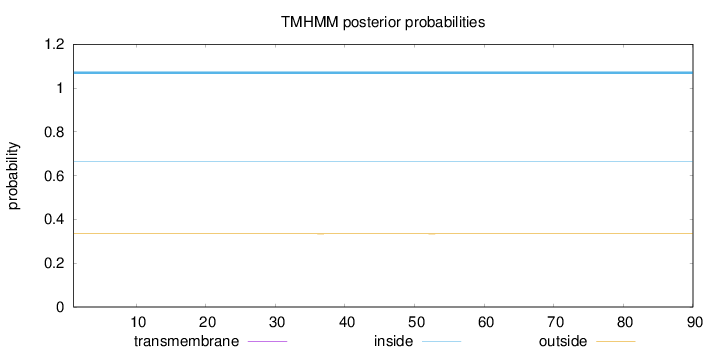
Length:
90
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00181
Exp number, first 60 AAs:
0.00181
Total prob of N-in:
0.66577
inside
1 - 90
Population Genetic Test Statistics
Pi
0.807891
Theta
5.142472
Tajima's D
-2.302381
CLR
1044.86631
CSRT
0.00199990000499975
Interpretation
Possibly Positive selection
