Gene
KWMTBOMO07324
Pre Gene Modal
BGIBMGA010622
Annotation
PREDICTED:_mediator_of_RNA_polymerase_II_transcription_subunit_8_[Papilio_polytes]
Full name
Mediator of RNA polymerase II transcription subunit 8
Alternative Name
Mediator complex subunit 8
Location in the cell
Mitochondrial Reliability : 1.6 Nuclear Reliability : 1.756
Sequence
CDS
ATGAATATGCAGCAGCGAGAAGAAAAGCAACTAGAAGCTACAATAGTTGCGATTTTAAATAGAGTAAATGATTTAAAAACTGCAATACAAGCATTGATAAATAAACTAGAAACGGAATACGAGACAATAAATTGGCCAACTTTTCTAGATAATTACGCGATTTTATCTGGTCACCTGACTGGCCTCAGTAAAATTCTCCAAGCCGAACAAGCAGCTTCATTGCGGTCTAGAATTGTTTTACCACTTCAATTAAGTTGCGAACGTGATGAAACACTGGCTCATTTGACAGAAGGCAGAGTTCCAGCTTGCACTCATGATTTAGTTCCAGATTTACTTCGTACCAAACCTGAACCCCAGGCTGAACAGCGTCTCCAGCAATTTAATCATAAAGCTAGTACTTTAAGCTATGATACAGCACAGAAACACATTGCTCAATTCACTAAAGTTGTCAGTCACGTTTGGGAAATTATATCCAAAGGCCGCGAAGACTGGGAAGGAGAGTCAATGAGATCTACAGGTATGCAACCAACACACAACATTGCTGACACGCATGCTTTAGTAACGGCAGTAGGAACGGGAAAAGGGCTACGACCAGGTATGGCGAATATGGTTGGTCAAGGGGCAGCTGGAATGGGACCAGCTCGAGGCCCGGCTCCACAACTCGGTCCTGCGCCCTCTACTCTTCCTAAGGCACCTTCAGCTATCAAAACCAATATCAAAGCTGCTAATCAAATACATCCTTACCAAAGATAA
Protein
MNMQQREEKQLEATIVAILNRVNDLKTAIQALINKLETEYETINWPTFLDNYAILSGHLTGLSKILQAEQAASLRSRIVLPLQLSCERDETLAHLTEGRVPACTHDLVPDLLRTKPEPQAEQRLQQFNHKASTLSYDTAQKHIAQFTKVVSHVWEIISKGREDWEGESMRSTGMQPTHNIADTHALVTAVGTGKGLRPGMANMVGQGAAGMGPARGPAPQLGPAPSTLPKAPSAIKTNIKAANQIHPYQR
Summary
Description
Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors.
Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors (By similarity). Required for activated transcription of the MtnA and MtnB genes.
Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors (By similarity).
Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors (By similarity). Required for activated transcription of the MtnA and MtnB genes.
Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors (By similarity).
Subunit
Component of the Mediator complex.
Similarity
Belongs to the Mediator complex subunit 8 family.
Keywords
Activator
Coiled coil
Complete proteome
Nucleus
Reference proteome
Transcription
Transcription regulation
Feature
chain Mediator of RNA polymerase II transcription subunit 8
Uniprot
H9JM69
A0A2A4J0A5
A0A2H1V5H4
A0A212ETY7
A0A194R2Q2
A0A3S2M5Q0
+ More
S4PT92 A0A0L7LIQ4 A0A194PGY0 A0A1E1WMH5 A0A1A9Y7U0 A0A1B0ALB6 A0A1A9ZQA0 D3TRA0 Q28Z80 B4GI55 A0A1A9UDX3 D6WPY4 A0A3B0JJY3 A0A310SIS4 A0A0L0CIH6 E0VXU9 A0A0L7R4Q7 A0A1W4VGL9 A0A067QSE2 A0A1W4WY44 A0A1W4WMB2 A0A1W4WYC6 A0A1W4WMD1 A0A1W4WY41 A0A1W4WX18 A0A0T6B3X9 A0A1I8PDB7 B3MHK9 A0A1L8DHR6 B4PAL9 B3NK78 B6IDR6 A1ZBT5 A0A1B0CD10 B4HQ65 B4QEN1 A0A154PK13 A0A1I8NDI3 A0A1B0GMN8 A0A2A3E126 A0A336LQP3 B4N5W5 A0A0J7L1F4 Q16RR7 F4X5G3 A0A151WYN6 A0A195F018 B4KN99 B4LMT0 A0A0K8WB67 A0A182ITE7 A0A182Y2V9 A0A088AD95 A0A0M4EGN9 A0A151IIT0 U5EYA0 A0A182JVK0 A0A034WT71 A0A232ELR0 A0A182WK07 V9IKH0 A0A182M7C5 A0A182P6E1 A0A182VCN2 A0A182UHS9 A0A182X6X0 A0A182KPR6 A0A182I214 Q7Q6D6 E2AUJ3 K7IQA2 A0A1Y1KQR8 A0A182NLN8 W8C353 A0A182H9G5 A0A026W041 A0A2M4BXX7 A0A182QEZ9 A0A158P2Z3 A0A084W2M7 A0A2M4AVN8 B4J7J2 N6TAE0 A0A0A9W294 A0A0A9W3F5 A0A023EL83 A0A1L8DFS7 W5JNG6 A0A3L8DRL3 E2C3H0 A0A1B6BX70 A0A069DQB9 A0A224XUS9 A0A023F9E2
S4PT92 A0A0L7LIQ4 A0A194PGY0 A0A1E1WMH5 A0A1A9Y7U0 A0A1B0ALB6 A0A1A9ZQA0 D3TRA0 Q28Z80 B4GI55 A0A1A9UDX3 D6WPY4 A0A3B0JJY3 A0A310SIS4 A0A0L0CIH6 E0VXU9 A0A0L7R4Q7 A0A1W4VGL9 A0A067QSE2 A0A1W4WY44 A0A1W4WMB2 A0A1W4WYC6 A0A1W4WMD1 A0A1W4WY41 A0A1W4WX18 A0A0T6B3X9 A0A1I8PDB7 B3MHK9 A0A1L8DHR6 B4PAL9 B3NK78 B6IDR6 A1ZBT5 A0A1B0CD10 B4HQ65 B4QEN1 A0A154PK13 A0A1I8NDI3 A0A1B0GMN8 A0A2A3E126 A0A336LQP3 B4N5W5 A0A0J7L1F4 Q16RR7 F4X5G3 A0A151WYN6 A0A195F018 B4KN99 B4LMT0 A0A0K8WB67 A0A182ITE7 A0A182Y2V9 A0A088AD95 A0A0M4EGN9 A0A151IIT0 U5EYA0 A0A182JVK0 A0A034WT71 A0A232ELR0 A0A182WK07 V9IKH0 A0A182M7C5 A0A182P6E1 A0A182VCN2 A0A182UHS9 A0A182X6X0 A0A182KPR6 A0A182I214 Q7Q6D6 E2AUJ3 K7IQA2 A0A1Y1KQR8 A0A182NLN8 W8C353 A0A182H9G5 A0A026W041 A0A2M4BXX7 A0A182QEZ9 A0A158P2Z3 A0A084W2M7 A0A2M4AVN8 B4J7J2 N6TAE0 A0A0A9W294 A0A0A9W3F5 A0A023EL83 A0A1L8DFS7 W5JNG6 A0A3L8DRL3 E2C3H0 A0A1B6BX70 A0A069DQB9 A0A224XUS9 A0A023F9E2
Pubmed
19121390
22118469
26354079
23622113
26227816
20353571
+ More
15632085 17994087 18362917 19820115 26108605 20566863 24845553 17550304 10731132 12537572 12537569 16751183 22936249 25315136 17510324 21719571 25244985 25348373 28648823 20966253 12364791 20798317 20075255 28004739 24495485 26483478 24508170 21347285 24438588 23537049 25401762 26823975 24945155 20920257 23761445 30249741 26334808 25474469
15632085 17994087 18362917 19820115 26108605 20566863 24845553 17550304 10731132 12537572 12537569 16751183 22936249 25315136 17510324 21719571 25244985 25348373 28648823 20966253 12364791 20798317 20075255 28004739 24495485 26483478 24508170 21347285 24438588 23537049 25401762 26823975 24945155 20920257 23761445 30249741 26334808 25474469
EMBL
BABH01033430
NWSH01004441
PCG65108.1
ODYU01000758
SOQ36026.1
AGBW02012509
+ More
OWR44965.1 KQ460845 KPJ12068.1 RSAL01000027 RVE51964.1 GAIX01012443 JAA80117.1 JTDY01000955 KOB75295.1 KQ459603 KPI92651.1 GDQN01002906 JAT88148.1 JXJN01030833 CCAG010004837 EZ423952 ADD20228.1 CM000071 EAL25734.1 CH479183 EDW36175.1 KQ971354 EFA06894.2 OUUW01000001 SPP72921.1 KQ762903 OAD55292.1 JRES01000349 KNC32007.1 AAZO01006138 DS235841 EEB18205.1 KQ414657 KOC65852.1 KK853011 KDR12538.1 LJIG01016021 KRT81851.1 CH902619 EDV35845.1 GFDF01008082 JAV06002.1 CM000158 EDW91410.1 CH954179 EDV55299.1 BT050506 ACJ13213.1 AE013599 AY071108 AJWK01007293 CH480816 EDW48690.1 CM000362 CM002911 EDX07905.1 KMY95252.1 KQ434924 KZC11560.1 AJVK01026357 KZ288467 PBC25400.1 UFQT01000048 SSX18983.1 CH964154 EDW79754.1 LBMM01001281 KMQ96587.1 CH477697 GL888716 EGI58319.1 KQ982649 KYQ52989.1 KQ981905 KYN33469.1 CH933808 EDW09952.1 CH940648 EDW61021.1 GDHF01012709 GDHF01003913 GDHF01000191 JAI39605.1 JAI48401.1 JAI52123.1 CP012524 ALC42751.1 KQ977459 KYN02576.1 GANO01000494 JAB59377.1 GAKP01001108 JAC57844.1 NNAY01003517 OXU19289.1 JR052037 AEY61613.1 AXCM01000897 AXCM01004559 APCN01000104 AAAB01008960 GL442829 EFN62887.1 GEZM01082928 JAV61217.1 GAMC01010312 GAMC01010311 JAB96244.1 JXUM01120630 KQ566414 KXJ70153.1 KK107526 EZA49270.1 GGFJ01008721 MBW57862.1 AXCN02002338 ADTU01007654 ATLV01019640 KE525275 KFB44471.1 GGFK01011538 MBW44859.1 CH916367 EDW02140.1 APGK01045744 KB741037 KB632173 ENN74703.1 ERL89549.1 GBHO01041660 GBRD01010087 GDHC01016594 GDHC01010044 GDHC01003061 JAG01944.1 JAG55737.1 JAQ02035.1 JAQ08585.1 JAQ15568.1 GBHO01041658 JAG01946.1 GAPW01003615 JAC09983.1 GFDF01008775 JAV05309.1 ADMH02000844 ETN64863.1 QOIP01000005 RLU23060.1 GL452320 EFN77508.1 GEDC01031425 GEDC01028738 GEDC01025722 GEDC01018758 GEDC01018500 GEDC01017339 GEDC01008340 GEDC01008050 GEDC01002178 GEDC01001730 GEDC01001606 JAS05873.1 JAS08560.1 JAS11576.1 JAS18540.1 JAS18798.1 JAS19959.1 JAS28958.1 JAS29248.1 JAS35120.1 JAS35568.1 JAS35692.1 GBGD01002626 JAC86263.1 GFTR01004174 JAW12252.1 GBBI01000712 JAC18000.1
OWR44965.1 KQ460845 KPJ12068.1 RSAL01000027 RVE51964.1 GAIX01012443 JAA80117.1 JTDY01000955 KOB75295.1 KQ459603 KPI92651.1 GDQN01002906 JAT88148.1 JXJN01030833 CCAG010004837 EZ423952 ADD20228.1 CM000071 EAL25734.1 CH479183 EDW36175.1 KQ971354 EFA06894.2 OUUW01000001 SPP72921.1 KQ762903 OAD55292.1 JRES01000349 KNC32007.1 AAZO01006138 DS235841 EEB18205.1 KQ414657 KOC65852.1 KK853011 KDR12538.1 LJIG01016021 KRT81851.1 CH902619 EDV35845.1 GFDF01008082 JAV06002.1 CM000158 EDW91410.1 CH954179 EDV55299.1 BT050506 ACJ13213.1 AE013599 AY071108 AJWK01007293 CH480816 EDW48690.1 CM000362 CM002911 EDX07905.1 KMY95252.1 KQ434924 KZC11560.1 AJVK01026357 KZ288467 PBC25400.1 UFQT01000048 SSX18983.1 CH964154 EDW79754.1 LBMM01001281 KMQ96587.1 CH477697 GL888716 EGI58319.1 KQ982649 KYQ52989.1 KQ981905 KYN33469.1 CH933808 EDW09952.1 CH940648 EDW61021.1 GDHF01012709 GDHF01003913 GDHF01000191 JAI39605.1 JAI48401.1 JAI52123.1 CP012524 ALC42751.1 KQ977459 KYN02576.1 GANO01000494 JAB59377.1 GAKP01001108 JAC57844.1 NNAY01003517 OXU19289.1 JR052037 AEY61613.1 AXCM01000897 AXCM01004559 APCN01000104 AAAB01008960 GL442829 EFN62887.1 GEZM01082928 JAV61217.1 GAMC01010312 GAMC01010311 JAB96244.1 JXUM01120630 KQ566414 KXJ70153.1 KK107526 EZA49270.1 GGFJ01008721 MBW57862.1 AXCN02002338 ADTU01007654 ATLV01019640 KE525275 KFB44471.1 GGFK01011538 MBW44859.1 CH916367 EDW02140.1 APGK01045744 KB741037 KB632173 ENN74703.1 ERL89549.1 GBHO01041660 GBRD01010087 GDHC01016594 GDHC01010044 GDHC01003061 JAG01944.1 JAG55737.1 JAQ02035.1 JAQ08585.1 JAQ15568.1 GBHO01041658 JAG01946.1 GAPW01003615 JAC09983.1 GFDF01008775 JAV05309.1 ADMH02000844 ETN64863.1 QOIP01000005 RLU23060.1 GL452320 EFN77508.1 GEDC01031425 GEDC01028738 GEDC01025722 GEDC01018758 GEDC01018500 GEDC01017339 GEDC01008340 GEDC01008050 GEDC01002178 GEDC01001730 GEDC01001606 JAS05873.1 JAS08560.1 JAS11576.1 JAS18540.1 JAS18798.1 JAS19959.1 JAS28958.1 JAS29248.1 JAS35120.1 JAS35568.1 JAS35692.1 GBGD01002626 JAC86263.1 GFTR01004174 JAW12252.1 GBBI01000712 JAC18000.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000283053
UP000037510
+ More
UP000053268 UP000092443 UP000092460 UP000092445 UP000092444 UP000001819 UP000008744 UP000078200 UP000007266 UP000268350 UP000037069 UP000009046 UP000053825 UP000192221 UP000027135 UP000192223 UP000095300 UP000007801 UP000002282 UP000008711 UP000000803 UP000092461 UP000001292 UP000000304 UP000076502 UP000095301 UP000092462 UP000242457 UP000007798 UP000036403 UP000008820 UP000007755 UP000075809 UP000078541 UP000009192 UP000008792 UP000075880 UP000076408 UP000005203 UP000092553 UP000078542 UP000075881 UP000215335 UP000075920 UP000075883 UP000075885 UP000075903 UP000075902 UP000076407 UP000075882 UP000075840 UP000007062 UP000000311 UP000002358 UP000075884 UP000069940 UP000249989 UP000053097 UP000075886 UP000005205 UP000030765 UP000001070 UP000019118 UP000030742 UP000000673 UP000279307 UP000008237
UP000053268 UP000092443 UP000092460 UP000092445 UP000092444 UP000001819 UP000008744 UP000078200 UP000007266 UP000268350 UP000037069 UP000009046 UP000053825 UP000192221 UP000027135 UP000192223 UP000095300 UP000007801 UP000002282 UP000008711 UP000000803 UP000092461 UP000001292 UP000000304 UP000076502 UP000095301 UP000092462 UP000242457 UP000007798 UP000036403 UP000008820 UP000007755 UP000075809 UP000078541 UP000009192 UP000008792 UP000075880 UP000076408 UP000005203 UP000092553 UP000078542 UP000075881 UP000215335 UP000075920 UP000075883 UP000075885 UP000075903 UP000075902 UP000076407 UP000075882 UP000075840 UP000007062 UP000000311 UP000002358 UP000075884 UP000069940 UP000249989 UP000053097 UP000075886 UP000005205 UP000030765 UP000001070 UP000019118 UP000030742 UP000000673 UP000279307 UP000008237
Pfam
PF10232 Med8
Interpro
IPR019364
Mediatior_Med8_fun/met
ProteinModelPortal
H9JM69
A0A2A4J0A5
A0A2H1V5H4
A0A212ETY7
A0A194R2Q2
A0A3S2M5Q0
+ More
S4PT92 A0A0L7LIQ4 A0A194PGY0 A0A1E1WMH5 A0A1A9Y7U0 A0A1B0ALB6 A0A1A9ZQA0 D3TRA0 Q28Z80 B4GI55 A0A1A9UDX3 D6WPY4 A0A3B0JJY3 A0A310SIS4 A0A0L0CIH6 E0VXU9 A0A0L7R4Q7 A0A1W4VGL9 A0A067QSE2 A0A1W4WY44 A0A1W4WMB2 A0A1W4WYC6 A0A1W4WMD1 A0A1W4WY41 A0A1W4WX18 A0A0T6B3X9 A0A1I8PDB7 B3MHK9 A0A1L8DHR6 B4PAL9 B3NK78 B6IDR6 A1ZBT5 A0A1B0CD10 B4HQ65 B4QEN1 A0A154PK13 A0A1I8NDI3 A0A1B0GMN8 A0A2A3E126 A0A336LQP3 B4N5W5 A0A0J7L1F4 Q16RR7 F4X5G3 A0A151WYN6 A0A195F018 B4KN99 B4LMT0 A0A0K8WB67 A0A182ITE7 A0A182Y2V9 A0A088AD95 A0A0M4EGN9 A0A151IIT0 U5EYA0 A0A182JVK0 A0A034WT71 A0A232ELR0 A0A182WK07 V9IKH0 A0A182M7C5 A0A182P6E1 A0A182VCN2 A0A182UHS9 A0A182X6X0 A0A182KPR6 A0A182I214 Q7Q6D6 E2AUJ3 K7IQA2 A0A1Y1KQR8 A0A182NLN8 W8C353 A0A182H9G5 A0A026W041 A0A2M4BXX7 A0A182QEZ9 A0A158P2Z3 A0A084W2M7 A0A2M4AVN8 B4J7J2 N6TAE0 A0A0A9W294 A0A0A9W3F5 A0A023EL83 A0A1L8DFS7 W5JNG6 A0A3L8DRL3 E2C3H0 A0A1B6BX70 A0A069DQB9 A0A224XUS9 A0A023F9E2
S4PT92 A0A0L7LIQ4 A0A194PGY0 A0A1E1WMH5 A0A1A9Y7U0 A0A1B0ALB6 A0A1A9ZQA0 D3TRA0 Q28Z80 B4GI55 A0A1A9UDX3 D6WPY4 A0A3B0JJY3 A0A310SIS4 A0A0L0CIH6 E0VXU9 A0A0L7R4Q7 A0A1W4VGL9 A0A067QSE2 A0A1W4WY44 A0A1W4WMB2 A0A1W4WYC6 A0A1W4WMD1 A0A1W4WY41 A0A1W4WX18 A0A0T6B3X9 A0A1I8PDB7 B3MHK9 A0A1L8DHR6 B4PAL9 B3NK78 B6IDR6 A1ZBT5 A0A1B0CD10 B4HQ65 B4QEN1 A0A154PK13 A0A1I8NDI3 A0A1B0GMN8 A0A2A3E126 A0A336LQP3 B4N5W5 A0A0J7L1F4 Q16RR7 F4X5G3 A0A151WYN6 A0A195F018 B4KN99 B4LMT0 A0A0K8WB67 A0A182ITE7 A0A182Y2V9 A0A088AD95 A0A0M4EGN9 A0A151IIT0 U5EYA0 A0A182JVK0 A0A034WT71 A0A232ELR0 A0A182WK07 V9IKH0 A0A182M7C5 A0A182P6E1 A0A182VCN2 A0A182UHS9 A0A182X6X0 A0A182KPR6 A0A182I214 Q7Q6D6 E2AUJ3 K7IQA2 A0A1Y1KQR8 A0A182NLN8 W8C353 A0A182H9G5 A0A026W041 A0A2M4BXX7 A0A182QEZ9 A0A158P2Z3 A0A084W2M7 A0A2M4AVN8 B4J7J2 N6TAE0 A0A0A9W294 A0A0A9W3F5 A0A023EL83 A0A1L8DFS7 W5JNG6 A0A3L8DRL3 E2C3H0 A0A1B6BX70 A0A069DQB9 A0A224XUS9 A0A023F9E2
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
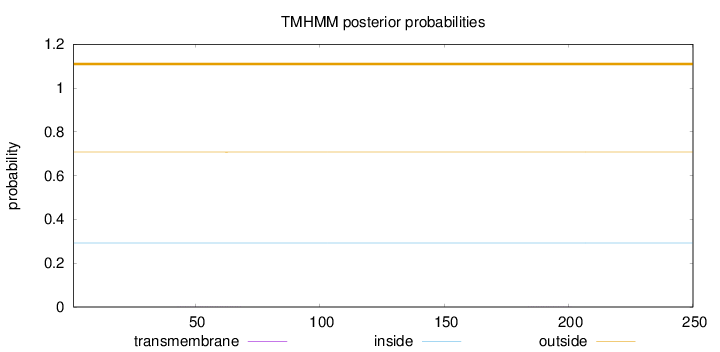
Length:
250
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01291
Exp number, first 60 AAs:
0.00653
Total prob of N-in:
0.29256
outside
1 - 250
Population Genetic Test Statistics
Pi
0.8784
Theta
5.621926
Tajima's D
-2.326997
CLR
332.663111
CSRT
0.00149992500374981
Interpretation
Possibly Positive selection
