Gene
KWMTBOMO07323 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010666
Annotation
chaperonin_subunit_4_delta_[Bombyx_mori]
Full name
T-complex protein 1 subunit delta
Alternative Name
CCT-delta
Location in the cell
Cytoplasmic Reliability : 2.65
Sequence
CDS
ATGGCTCCGAAAGCTGGCGGTGATGCTATTAAAGCTAATTCTTCAGTATATAAAGATAAAAGTAAACCTACAGACATCAGACTGAGCAACATAAATGCCGCAAAAGCTGTTGCTGATGCCATCCGCACAAGCTTAGGGCCTCGTGGAATGGATAAGATGATTCAAGCAGCCAATGGAGAAGTTACAATAACTAATGATGGAGCAACAATTTTGAAACAAATGAGTGTTATTCATCCAGCTGCAAAAATGTTGGTAGAATTATCTCGTGCTCAAGATATTGAAGCTGGTGATGGAACAACATCAGTAGTTGTTATTGCTGGTGCGTTGTTGGATTCTGCTGAGAAACTTCTTCAGAAAGGCATACATCCAACTGTAATTTCTGATGGTTTCCAAAAAGCCCTTCAATTGGCTTTACAGGTGGTCGAAAATATGTCAACTCCAGTGGACTTGAATAATGAAGATGCTTTGCTCAAGGCAGCTGCAACATCACTAAATTCCAAAGTAGTTTCACAACACTCAACTATTTTGGCACCCATTGCAGTGCAAGCAATTCGAGCAGTAATGGAACCTATTGTTAGTGGTGTTGGTGCAAGAGTTGACCTCAGAGATGTGAAAGTCATTGAGCGTATAGGTGGAACTGTAGAAGATGCTGAGCTAATCCAAGGATTGGTCATTCCGCATCGTGCAGCCAATGTGAATGGCCCTCATCGCATTGAAAAGGCTAAAGTTGGTCTTATACAATTTTGCATTTCGCCTCCCAAAACTGATATGGACCACAATGTTATTGTATCAGATTATGCAGCAATGGATCGTGTGTTGAAGGAAGAACGCCAATATATTTTGAATATTGTAAAACAGATCAAAAAAGCTGGTTGTAATGTACTTTTAGTTCAAAAATCTATATTAAGAGATGCCCTGAGTGACTTGGCTGTACATTTCTTAGACAAAATTAAAACAATGGTCATTAAAGATATTGAACGTGAAGACATTGAATTTGTTTGCAAAACCTTAGGATGCAGGCCAATTGCTTCTTTGGATCATTTTACAGCTGAAAATTTAGTCAATGTTGATTTGGTTGAAGAAATAAATGAACCTGGTGGAAAATATATAAAGATGACGGGTTGCGGTCAGGGCGGTCGCACAGTGAGCGTATTGGTTCGCGGATCGAGTGGCGCCGTGGTGGCAGAGGCAGCGCGATCTCTTCACGACGCCTTATGTGCCGTGCGCTGTGTGGCGCGCCGCCCTGCATTGTTGGCCGGCGGCGGCGCTCCCGAGGCTGCAGCGGCGGCCAGTCTCGCGCGTGCTTCTCTCGCAGCACCCGGAGCCGATCACTATTGTCTACGTGCCTACGCCGATGCTCTCGAAGTCATACCCTCCACGCTTGCCGAGAATGCAGGTTTGAACCCAATTGAGACCGTAACTGAGCTACGAGCCGCTCACGCTGCTGGCGCTGGCGGTGGCGCCGGTGTCAACGTACGCCGCGGACGAGTTACAGATATGCGCACGGAGCATGTGCTGCAACCGCTTCATGTTACAGCTTCCGCGCTCGCACTCGCAACCGAAACTGTGCGTGCTATACTCAAAATTGATGACATTATAAATACAATGAATTAG
Protein
MAPKAGGDAIKANSSVYKDKSKPTDIRLSNINAAKAVADAIRTSLGPRGMDKMIQAANGEVTITNDGATILKQMSVIHPAAKMLVELSRAQDIEAGDGTTSVVVIAGALLDSAEKLLQKGIHPTVISDGFQKALQLALQVVENMSTPVDLNNEDALLKAAATSLNSKVVSQHSTILAPIAVQAIRAVMEPIVSGVGARVDLRDVKVIERIGGTVEDAELIQGLVIPHRAANVNGPHRIEKAKVGLIQFCISPPKTDMDHNVIVSDYAAMDRVLKEERQYILNIVKQIKKAGCNVLLVQKSILRDALSDLAVHFLDKIKTMVIKDIEREDIEFVCKTLGCRPIASLDHFTAENLVNVDLVEEINEPGGKYIKMTGCGQGGRTVSVLVRGSSGAVVAEAARSLHDALCAVRCVARRPALLAGGGAPEAAAAASLARASLAAPGADHYCLRAYADALEVIPSTLAENAGLNPIETVTELRAAHAAGAGGGAGVNVRRGRVTDMRTEHVLQPLHVTASALALATETVRAILKIDDIINTMN
Summary
Description
Molecular chaperone; assists the folding of proteins upon ATP hydrolysis. Known to play a role, in vitro, in the folding of actin and tubulin (By similarity).
Subunit
Heterooligomeric complex of about 850 to 900 kDa that forms two stacked rings, 12 to 16 nm in diameter.
Similarity
Belongs to the TCP-1 chaperonin family.
Keywords
ATP-binding
Chaperone
Cytoplasm
Nucleotide-binding
Feature
chain T-complex protein 1 subunit delta
Uniprot
Q2F6C4
H9JMB3
A0A2H1V5C0
A0A212ETZ6
S4PHL1
A0A194PH72
+ More
A0A194R301 A0A2A4IYU0 B4MDL7 A0A1Y1M3T7 B4MWN7 B4KIN7 V5G4G3 B3MNJ7 B4JCN1 A0A3B0JML9 A0A0K8UHH1 K7ILV5 Q29L81 W8APG2 A0A034WRN7 A0A0C9QQQ2 B4HB36 D2A1X9 A0A0L0CII4 A0A1I8MLK1 A0A0N0BKX4 A0A1B6LZ41 T1PFX8 Q9VK69 A0A1B6EX07 A0A182LWT3 A0A1A9WUI1 B4P2H9 A0A2A3E687 V9IHB3 A0A088AHM7 A0A0J9R146 A0A0M4E5N0 B3N3D6 A0A1B6DNB6 A0A182VUY3 A0A1W4V5I8 A0A097P6J3 A0A1I8NV30 B4IEB2 W5J975 A0A154P1G3 A0A2M4A9F9 A0A310SHQ9 A0A084VPG9 A0A182S4R7 A0A2M4BJP8 A0A2M4BK72 A0A182PD84 A0A026W822 A0A1L8ECW3 A0A2M3Z334 A0A182F559 A0A232FL45 E9IGN8 A0A0M4E1N5 Q171B7 A0A182JIF7 E2AX57 A0A182TXH4 A0A182X7L7 Q7QG50 A0A182IE50 A0A182QCT9 A0A1Q3F3R6 A0A182K0N8 E2B8Q4 A0A151X3S1 A0A182VJ93 A0A182Y5V1 A0A1L8DUD3 A0A1L8DUB8 Q9NB32 A0A195EEX5 A0A0J7L083 A0A1W4X1L7 A0A195ESL8 F4WQT6 A0A182GEL1 A0A1S3D225 A0A023EWC4 A0A067RDC2 A0A158NRS2 A0A023ETP0 W5U591 B0W5S5 A0A336KPC9 A0A0K8TST0 A0A195BRM7 U5EX42 A0A195D142 A0A224XN34 A0A0A9Y6Y1 J9JSI4
A0A194R301 A0A2A4IYU0 B4MDL7 A0A1Y1M3T7 B4MWN7 B4KIN7 V5G4G3 B3MNJ7 B4JCN1 A0A3B0JML9 A0A0K8UHH1 K7ILV5 Q29L81 W8APG2 A0A034WRN7 A0A0C9QQQ2 B4HB36 D2A1X9 A0A0L0CII4 A0A1I8MLK1 A0A0N0BKX4 A0A1B6LZ41 T1PFX8 Q9VK69 A0A1B6EX07 A0A182LWT3 A0A1A9WUI1 B4P2H9 A0A2A3E687 V9IHB3 A0A088AHM7 A0A0J9R146 A0A0M4E5N0 B3N3D6 A0A1B6DNB6 A0A182VUY3 A0A1W4V5I8 A0A097P6J3 A0A1I8NV30 B4IEB2 W5J975 A0A154P1G3 A0A2M4A9F9 A0A310SHQ9 A0A084VPG9 A0A182S4R7 A0A2M4BJP8 A0A2M4BK72 A0A182PD84 A0A026W822 A0A1L8ECW3 A0A2M3Z334 A0A182F559 A0A232FL45 E9IGN8 A0A0M4E1N5 Q171B7 A0A182JIF7 E2AX57 A0A182TXH4 A0A182X7L7 Q7QG50 A0A182IE50 A0A182QCT9 A0A1Q3F3R6 A0A182K0N8 E2B8Q4 A0A151X3S1 A0A182VJ93 A0A182Y5V1 A0A1L8DUD3 A0A1L8DUB8 Q9NB32 A0A195EEX5 A0A0J7L083 A0A1W4X1L7 A0A195ESL8 F4WQT6 A0A182GEL1 A0A1S3D225 A0A023EWC4 A0A067RDC2 A0A158NRS2 A0A023ETP0 W5U591 B0W5S5 A0A336KPC9 A0A0K8TST0 A0A195BRM7 U5EX42 A0A195D142 A0A224XN34 A0A0A9Y6Y1 J9JSI4
Pubmed
19121390
22118469
23622113
26354079
17994087
28004739
+ More
20075255 15632085 24495485 25348373 18362917 19820115 26108605 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 20920257 23761445 24438588 24508170 30249741 28648823 21282665 17510324 20798317 12364791 14747013 17210077 25244985 11762197 21719571 26483478 24945155 24845553 21347285 26369729 25401762 26823975
20075255 15632085 24495485 25348373 18362917 19820115 26108605 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 20920257 23761445 24438588 24508170 30249741 28648823 21282665 17510324 20798317 12364791 14747013 17210077 25244985 11762197 21719571 26483478 24945155 24845553 21347285 26369729 25401762 26823975
EMBL
DQ311148
ABD36093.1
BABH01033429
BABH01033430
ODYU01000758
SOQ36027.1
+ More
AGBW02012509 OWR44966.1 GAIX01003232 JAA89328.1 KQ459603 KPI92652.1 KQ460845 KPJ12067.1 NWSH01004441 PCG65107.1 CH940661 EDW71278.1 GEZM01045202 JAV77867.1 CH963857 EDW76526.1 CH933807 EDW11380.1 GALX01003551 JAB64915.1 CH902620 EDV32105.1 CH916368 EDW04195.1 OUUW01000006 SPP82103.1 GDHF01026177 JAI26137.1 CH379061 EAL32943.1 GAMC01020147 JAB86408.1 GAKP01001658 JAC57294.1 GBYB01002992 JAG72759.1 CH479247 EDW37839.1 KQ971338 EFA02141.1 JRES01000438 KNC31299.1 KQ435689 KOX81218.1 GEBQ01011020 JAT28957.1 KA647661 AFP62290.1 AE014134 AY128484 AAF53210.1 AAM75077.1 GECZ01027270 GECZ01026922 JAS42499.1 JAS42847.1 AXCM01006443 CM000157 EDW88209.1 KZ288371 PBC26736.1 JR042985 AEY59704.1 CM002910 KMY89947.1 CP012523 ALC39446.1 CH954177 EDV58776.1 GEDC01010133 JAS27165.1 KM215142 AIU39615.1 CH480831 EDW45939.1 ADMH02002089 ETN59400.1 KQ434796 KZC05662.1 GGFK01004088 MBW37409.1 KQ768478 OAD53120.1 ATLV01015004 KE524999 KFB39863.1 GGFJ01004158 MBW53299.1 GGFJ01004263 MBW53404.1 KK107347 QOIP01000007 EZA52257.1 RLU21000.1 GFDG01002380 JAV16419.1 GGFM01002152 MBW22903.1 NNAY01000091 OXU31067.1 GL763054 EFZ20189.1 ALC39328.1 CH477460 EAT40572.1 GL443520 EFN62007.1 AAAB01008839 EAA05907.2 APCN01004564 AXCN02000859 GFDL01012868 JAV22177.1 GL446361 EFN87960.1 KQ982562 KYQ54920.1 GFDF01004045 JAV10039.1 GFDF01004046 JAV10038.1 AF271209 KQ978983 KYN26825.1 LBMM01001595 KMQ96041.1 KQ981993 KYN30907.1 GL888275 EGI63449.1 JXUM01058230 JXUM01058231 JXUM01058232 KQ561993 KXJ76960.1 GAPW01000894 JAC12704.1 KK852777 KDR16778.1 ADTU01024286 GAPW01000895 JAC12703.1 KF813101 AHH35014.1 DS231844 EDS35728.1 UFQS01000672 UFQT01000055 UFQT01000672 SSX06011.1 SSX19079.1 GDAI01000415 JAI17188.1 KQ976424 KYM88596.1 GANO01001132 JAB58739.1 KQ977041 KYN06124.1 GFTR01006995 JAW09431.1 GBHO01016761 GBRD01010602 GDHC01007949 JAG26843.1 JAG55222.1 JAQ10680.1 ABLF02036421
AGBW02012509 OWR44966.1 GAIX01003232 JAA89328.1 KQ459603 KPI92652.1 KQ460845 KPJ12067.1 NWSH01004441 PCG65107.1 CH940661 EDW71278.1 GEZM01045202 JAV77867.1 CH963857 EDW76526.1 CH933807 EDW11380.1 GALX01003551 JAB64915.1 CH902620 EDV32105.1 CH916368 EDW04195.1 OUUW01000006 SPP82103.1 GDHF01026177 JAI26137.1 CH379061 EAL32943.1 GAMC01020147 JAB86408.1 GAKP01001658 JAC57294.1 GBYB01002992 JAG72759.1 CH479247 EDW37839.1 KQ971338 EFA02141.1 JRES01000438 KNC31299.1 KQ435689 KOX81218.1 GEBQ01011020 JAT28957.1 KA647661 AFP62290.1 AE014134 AY128484 AAF53210.1 AAM75077.1 GECZ01027270 GECZ01026922 JAS42499.1 JAS42847.1 AXCM01006443 CM000157 EDW88209.1 KZ288371 PBC26736.1 JR042985 AEY59704.1 CM002910 KMY89947.1 CP012523 ALC39446.1 CH954177 EDV58776.1 GEDC01010133 JAS27165.1 KM215142 AIU39615.1 CH480831 EDW45939.1 ADMH02002089 ETN59400.1 KQ434796 KZC05662.1 GGFK01004088 MBW37409.1 KQ768478 OAD53120.1 ATLV01015004 KE524999 KFB39863.1 GGFJ01004158 MBW53299.1 GGFJ01004263 MBW53404.1 KK107347 QOIP01000007 EZA52257.1 RLU21000.1 GFDG01002380 JAV16419.1 GGFM01002152 MBW22903.1 NNAY01000091 OXU31067.1 GL763054 EFZ20189.1 ALC39328.1 CH477460 EAT40572.1 GL443520 EFN62007.1 AAAB01008839 EAA05907.2 APCN01004564 AXCN02000859 GFDL01012868 JAV22177.1 GL446361 EFN87960.1 KQ982562 KYQ54920.1 GFDF01004045 JAV10039.1 GFDF01004046 JAV10038.1 AF271209 KQ978983 KYN26825.1 LBMM01001595 KMQ96041.1 KQ981993 KYN30907.1 GL888275 EGI63449.1 JXUM01058230 JXUM01058231 JXUM01058232 KQ561993 KXJ76960.1 GAPW01000894 JAC12704.1 KK852777 KDR16778.1 ADTU01024286 GAPW01000895 JAC12703.1 KF813101 AHH35014.1 DS231844 EDS35728.1 UFQS01000672 UFQT01000055 UFQT01000672 SSX06011.1 SSX19079.1 GDAI01000415 JAI17188.1 KQ976424 KYM88596.1 GANO01001132 JAB58739.1 KQ977041 KYN06124.1 GFTR01006995 JAW09431.1 GBHO01016761 GBRD01010602 GDHC01007949 JAG26843.1 JAG55222.1 JAQ10680.1 ABLF02036421
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000218220
UP000008792
+ More
UP000007798 UP000009192 UP000007801 UP000001070 UP000268350 UP000002358 UP000001819 UP000008744 UP000007266 UP000037069 UP000095301 UP000053105 UP000000803 UP000075883 UP000091820 UP000002282 UP000242457 UP000005203 UP000092553 UP000008711 UP000075920 UP000192221 UP000095300 UP000001292 UP000000673 UP000076502 UP000030765 UP000075900 UP000075885 UP000053097 UP000279307 UP000069272 UP000215335 UP000008820 UP000075880 UP000000311 UP000075902 UP000076407 UP000007062 UP000075840 UP000075886 UP000075881 UP000008237 UP000075809 UP000075903 UP000076408 UP000078492 UP000036403 UP000192223 UP000078541 UP000007755 UP000069940 UP000249989 UP000079169 UP000027135 UP000005205 UP000002320 UP000078540 UP000078542 UP000007819
UP000007798 UP000009192 UP000007801 UP000001070 UP000268350 UP000002358 UP000001819 UP000008744 UP000007266 UP000037069 UP000095301 UP000053105 UP000000803 UP000075883 UP000091820 UP000002282 UP000242457 UP000005203 UP000092553 UP000008711 UP000075920 UP000192221 UP000095300 UP000001292 UP000000673 UP000076502 UP000030765 UP000075900 UP000075885 UP000053097 UP000279307 UP000069272 UP000215335 UP000008820 UP000075880 UP000000311 UP000075902 UP000076407 UP000007062 UP000075840 UP000075886 UP000075881 UP000008237 UP000075809 UP000075903 UP000076408 UP000078492 UP000036403 UP000192223 UP000078541 UP000007755 UP000069940 UP000249989 UP000079169 UP000027135 UP000005205 UP000002320 UP000078540 UP000078542 UP000007819
PRIDE
Pfam
PF00118 Cpn60_TCP1
Interpro
Gene 3D
ProteinModelPortal
Q2F6C4
H9JMB3
A0A2H1V5C0
A0A212ETZ6
S4PHL1
A0A194PH72
+ More
A0A194R301 A0A2A4IYU0 B4MDL7 A0A1Y1M3T7 B4MWN7 B4KIN7 V5G4G3 B3MNJ7 B4JCN1 A0A3B0JML9 A0A0K8UHH1 K7ILV5 Q29L81 W8APG2 A0A034WRN7 A0A0C9QQQ2 B4HB36 D2A1X9 A0A0L0CII4 A0A1I8MLK1 A0A0N0BKX4 A0A1B6LZ41 T1PFX8 Q9VK69 A0A1B6EX07 A0A182LWT3 A0A1A9WUI1 B4P2H9 A0A2A3E687 V9IHB3 A0A088AHM7 A0A0J9R146 A0A0M4E5N0 B3N3D6 A0A1B6DNB6 A0A182VUY3 A0A1W4V5I8 A0A097P6J3 A0A1I8NV30 B4IEB2 W5J975 A0A154P1G3 A0A2M4A9F9 A0A310SHQ9 A0A084VPG9 A0A182S4R7 A0A2M4BJP8 A0A2M4BK72 A0A182PD84 A0A026W822 A0A1L8ECW3 A0A2M3Z334 A0A182F559 A0A232FL45 E9IGN8 A0A0M4E1N5 Q171B7 A0A182JIF7 E2AX57 A0A182TXH4 A0A182X7L7 Q7QG50 A0A182IE50 A0A182QCT9 A0A1Q3F3R6 A0A182K0N8 E2B8Q4 A0A151X3S1 A0A182VJ93 A0A182Y5V1 A0A1L8DUD3 A0A1L8DUB8 Q9NB32 A0A195EEX5 A0A0J7L083 A0A1W4X1L7 A0A195ESL8 F4WQT6 A0A182GEL1 A0A1S3D225 A0A023EWC4 A0A067RDC2 A0A158NRS2 A0A023ETP0 W5U591 B0W5S5 A0A336KPC9 A0A0K8TST0 A0A195BRM7 U5EX42 A0A195D142 A0A224XN34 A0A0A9Y6Y1 J9JSI4
A0A194R301 A0A2A4IYU0 B4MDL7 A0A1Y1M3T7 B4MWN7 B4KIN7 V5G4G3 B3MNJ7 B4JCN1 A0A3B0JML9 A0A0K8UHH1 K7ILV5 Q29L81 W8APG2 A0A034WRN7 A0A0C9QQQ2 B4HB36 D2A1X9 A0A0L0CII4 A0A1I8MLK1 A0A0N0BKX4 A0A1B6LZ41 T1PFX8 Q9VK69 A0A1B6EX07 A0A182LWT3 A0A1A9WUI1 B4P2H9 A0A2A3E687 V9IHB3 A0A088AHM7 A0A0J9R146 A0A0M4E5N0 B3N3D6 A0A1B6DNB6 A0A182VUY3 A0A1W4V5I8 A0A097P6J3 A0A1I8NV30 B4IEB2 W5J975 A0A154P1G3 A0A2M4A9F9 A0A310SHQ9 A0A084VPG9 A0A182S4R7 A0A2M4BJP8 A0A2M4BK72 A0A182PD84 A0A026W822 A0A1L8ECW3 A0A2M3Z334 A0A182F559 A0A232FL45 E9IGN8 A0A0M4E1N5 Q171B7 A0A182JIF7 E2AX57 A0A182TXH4 A0A182X7L7 Q7QG50 A0A182IE50 A0A182QCT9 A0A1Q3F3R6 A0A182K0N8 E2B8Q4 A0A151X3S1 A0A182VJ93 A0A182Y5V1 A0A1L8DUD3 A0A1L8DUB8 Q9NB32 A0A195EEX5 A0A0J7L083 A0A1W4X1L7 A0A195ESL8 F4WQT6 A0A182GEL1 A0A1S3D225 A0A023EWC4 A0A067RDC2 A0A158NRS2 A0A023ETP0 W5U591 B0W5S5 A0A336KPC9 A0A0K8TST0 A0A195BRM7 U5EX42 A0A195D142 A0A224XN34 A0A0A9Y6Y1 J9JSI4
PDB
4B2T
E-value=3.33947e-165,
Score=1494
Ontologies
GO
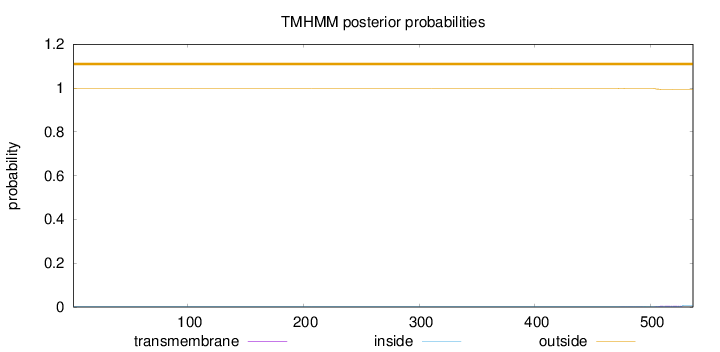
Topology
Subcellular location
Cytoplasm
Length:
537
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.12433
Exp number, first 60 AAs:
0.0006
Total prob of N-in:
0.00120
outside
1 - 537
Population Genetic Test Statistics
Pi
5.277103
Theta
12.754298
Tajima's D
-1.645516
CLR
3.674301
CSRT
0.0430478476076196
Interpretation
Uncertain
