Gene
KWMTBOMO07322
Pre Gene Modal
BGIBMGA010665
Annotation
hypothetical_protein_KGM_15744_[Danaus_plexippus]
Full name
DNA ligase
+ More
Rab3 GTPase-activating protein regulatory subunit
Rab3 GTPase-activating protein regulatory subunit
Location in the cell
PlasmaMembrane Reliability : 2.987
Sequence
CDS
ATGTCTTGTCAAATAGAACAAATTGCTGACTGTGGTGATTTATCTAGTTTAGCACGAGCACTGTTTTTAAGCAGTAATGAAGAACAAAATTGGTTGTCCCAATGTTGTGTTTCTCTATCATCTTGTGGTAACTTGCTTGCTGTTGGCTATCGCAATCGTCTTTGTATACTCACCACACAATGGATTAGTTCGACACAAAGCAATACTTTTTTGATATCTTGGAGTGGAACTTTACCTGCAGATATATCAACAGTATTGGTATTGCCAATTTGCCCATCTCATCATTCTTTCCAGAATGGCCCGGACTGGTTTTGCATTGTAGTTGGATTCAAAAACGGAAACATTGCTTTCCATACAAATACAGGCCATTTATTATTATCTGAAAAAATTGATGAAAAACCAGTATTGAAAATATCTTGTCATACTGGAACATATGGCACATTACCTGATGATGTTCATGTATTGTTTCATACTTGTGAATGTATTATATCAGGAGCAACGCTATTTCCAACACTGCGAAACGCAAAGGCACAATTAGCAAAAGTGCAAGCTGGTATTCAAGACTATTATAGTGTGGATAATCGTAGCATTAGCATAAAGAAATGGTTGTATACTGAACAAGAAGTTGTAAATGATGCTGCAGTTGTGGGATTAGAATTAAAAAATGCCTACGATCATTTACTAGCAGCTTCAACATATGGTGGTTATGATGTATGGTACCGTTCAGTCCCTCCAGTGAATACGTTAATATTAGGAGCTGGGCAGAAACCATATGTAGGTTTCCATTATGCTCTGGAGGGTGGCACTGCGCCCCCTTTACAAGATGTAGCCAGAGCTGTTGCACATAAAATCAAATCTGCTCTACCAGGCTGGCTAGGCGGGGCCTCGACAGAGGGCAGTGTGAGCGCGGCCGAGCCACGGATACGTTCGGAGCCCCTGTCCCTGCGCAGCGGCCTGCACGACGCGCAGAGGCACGGCACCTCCGTCACCGTCTCGCCGGACCGCCGCCTGGCTGCGGTCGTCGACAGCTTCGGCCGAGTCGCCGTACTGGACGTTAACAGAGGACATCTAATTAGACTAATAAAAGGTTGCAGAGACGCACAGTGTGCATTCGTCCAAATTTTTGAATTAGATAACAAGAAACCTCAATTATCTTTTGTTAAAGAAATAAGAAGGGCAATTTTTTTAATTATCTATAATCCCAAGAAAGGATTAATTGATATTCGTCTAATGCAAAGAGGGAACAGAGTAGCCGTTTTTACGGCTACGAAAAACGGCCAACTTTTATACAATACTTGTGGTCTAGTTGGAGCTGATAAAAATTATACCCAAAAAAAACTAAATTTACCAGAATTTCAGTGTGTTTTATTAGATCCAGATGGCAAGCTCAAAAAATTCAGTATTCCATTTTTTTACGCATTAGATGGTGTACATTCCGAAAGATCGAAAGATTTGAACACTTTGAAAGAATTACGTGATCTCATTAAAAACAATACAAGTAACTTTGAAGATTATGAGCAAAAGCTATTTAGTAAAGTATCCGAAATTAAAACATTAGAATTAAAGAAACATTTTCTAGAAATGCTCATTAGAAATTATGATACGTCTCCAACAATTATTATCTCATGTTTGCAAATATTCTGGGATAGCACAAAAGAGGAATCCGTGAGCAGTCAGCAACGTGAAACTCAACACTATTTTGCCAATCTAGGTTTGCTTACTATATTTTATATGTGCATAAATGACGATTGTGAAAGAGATGAAATAGAATTTGCAACTGAATTGCGTAATTTCCTTGGCTTAGAGGTAAATGAAAGTTGTTTGGAAAAAACAATAGACAACCAGAATGAAGACAACAGTTTGTTTCATCTACTAGAGGATGACAGTTGTATTTTGGAAAGATTACTGGTTTTAGCACACGAAAACAATTATAAACACCACGCACGGGTAACTTTTGCCGATACAAGTATGACTACCTATAAAAAATTCATTTCCTGTTTTAATTTAGAAGAAAAATCTAAATATATATCCTTAAAGGCCAATGCTTCGGTGGACAAGCTAAATAACTTAGCCACAATAACGTTCACGTCAATATTCAAATTAAAAAACTTTTCAATGTTGGCTAGTTATGTCAACAAATGCAATATCGACCCGAAAGAGATTGTGAAGCTTTTAATAACACATTTCATGAATATGTCTTTGGAAGAGATAACCATTGACTTAGTCGAGAAATGGACAATGGCCGTATACCACGTGTGTACGGTCACAAAAGACGCTACTAATATTATTTATAATGAAATTTCTCATTGGTGGGAAGACATCCGGAACGTTTTGGTTGAAATGCCTTGTCCCTTACGAAGCATGATAGTTGCAATGGCTTGTAAAGTAATCAGTAGGATTTTGGAAAGTAACAATGCTGAAAGCGACGATGTGTGGGAATCTGTGACCAAAGAAAACGCCAAATGGGGTATTTTAATTGGAAAACTTGAAGATATTTCTATACTCAGTATCGTATTAATGTGTAAAGACACATTTAATGGCAATTCTCTTCCTTCCCTGGAAATGGATGATTTCAATATCAACCTAAAATACATTTATACAAGGGGAAAGGGATCAATTACTGAACTGATTGCCAAGTGGCTATGCAGCATGGGAGCGACTCCTGCGGCGATAGTGGCTAACGAGCTAATGGAAACGATAGCAGCTAACTCTGAACTAACTGATGAGGACGACAATGATGACAACACCACTGATCTCCTGTTTGTTGAAAATAATCGTTATGCCGTTGACGAAAATACAAGAATATTTAAATGGCTCTCGCTTCTGCGCCGTCAATTCCCTCTCAGTACGTCCGCAAATTATATAATTACCAATATGTGTTGGGAATACGCAATGGCATGGCAGAAATGTATGAAGAATACATTGCTTCTTGATGCTGTTATTCACTGTCTTTCTAGTATATCTGACATTCATATACGGTTAGGGTTGTTTTCAATTATTTGGGCAACCTACATCAAACAAATATTTGAAATTAGTTGCCGTTTAGTTAACAAAGTCGGACGTTTGCCAAAAGACCCTCTTTGCTTACAAGATATCAGTTTCGACAATACAACCTTAATCAACTTCTTCCGTTTAACAACATTGTACTTGAATGATTTTCAAAATTGTTTGAAACTGTCATACAGTCATAATAAAAAGAAGGTGGAATTTGAAAAGATTTGGGAAGAAAGTATGCCTTCTTTAGTTGAAGTAACACAACACACAAAGAATGTAAACGATTGTATATTAACTTTAAATTACCAAGTATCTTGCACTCTGTACTATCAATGTCACTTCAATATCAAATTGTCGAAACCCCTCGATGTATTGTTCGACATCGACTATCATTATATTCTGGAAGCACTTACTGGTAATGTTGTGAATAGAGAAATTAATATGAAACCGTCTGAAAAACTCCGGAATCCACGAATGAAGTTTCTAACCAAACTTATAAGAGCATCTGTAGAAACTATAAGTTGTTCTGATGGTCACGAATCATACAGAACATATGATAGTGGTGACTGTCAAATGTGGATTGAAAAAATTTCCATTCTAGCGAACCTGTGGAGCATAGACGACGAATTCATAAAGAGGCAGCAGGTAATCGCTTTATATCACGTGGGTTACGATACACTAGCAGAAAATACAATGAACTCGATCCGTAATCCTGTGCCAGTGCTTGCTCCAATTTTGGCTATTACGATCCAACGTCTTAAACGAAGCCTAGAACATTCTAATAATCAAGCCGAATGGATTGTCAGTATTCCACCACAACTCTACAAACGTTTGCAAAATACTGTCCTGGACACATCAATTCCAGCACATCCGTCACTTTCCACCACAATATTAGTACTCCAAAAGTTGCTGTTTCAATTAAACAAAAAATCTCCTGATCCGGCTCATCTTCAGGATATAAAATTAATAGAGCTGATTATAGAAAACTGTGAAATTATTATTAGAAGAAAATTGTAA
Protein
MSCQIEQIADCGDLSSLARALFLSSNEEQNWLSQCCVSLSSCGNLLAVGYRNRLCILTTQWISSTQSNTFLISWSGTLPADISTVLVLPICPSHHSFQNGPDWFCIVVGFKNGNIAFHTNTGHLLLSEKIDEKPVLKISCHTGTYGTLPDDVHVLFHTCECIISGATLFPTLRNAKAQLAKVQAGIQDYYSVDNRSISIKKWLYTEQEVVNDAAVVGLELKNAYDHLLAASTYGGYDVWYRSVPPVNTLILGAGQKPYVGFHYALEGGTAPPLQDVARAVAHKIKSALPGWLGGASTEGSVSAAEPRIRSEPLSLRSGLHDAQRHGTSVTVSPDRRLAAVVDSFGRVAVLDVNRGHLIRLIKGCRDAQCAFVQIFELDNKKPQLSFVKEIRRAIFLIIYNPKKGLIDIRLMQRGNRVAVFTATKNGQLLYNTCGLVGADKNYTQKKLNLPEFQCVLLDPDGKLKKFSIPFFYALDGVHSERSKDLNTLKELRDLIKNNTSNFEDYEQKLFSKVSEIKTLELKKHFLEMLIRNYDTSPTIIISCLQIFWDSTKEESVSSQQRETQHYFANLGLLTIFYMCINDDCERDEIEFATELRNFLGLEVNESCLEKTIDNQNEDNSLFHLLEDDSCILERLLVLAHENNYKHHARVTFADTSMTTYKKFISCFNLEEKSKYISLKANASVDKLNNLATITFTSIFKLKNFSMLASYVNKCNIDPKEIVKLLITHFMNMSLEEITIDLVEKWTMAVYHVCTVTKDATNIIYNEISHWWEDIRNVLVEMPCPLRSMIVAMACKVISRILESNNAESDDVWESVTKENAKWGILIGKLEDISILSIVLMCKDTFNGNSLPSLEMDDFNINLKYIYTRGKGSITELIAKWLCSMGATPAAIVANELMETIAANSELTDEDDNDDNTTDLLFVENNRYAVDENTRIFKWLSLLRRQFPLSTSANYIITNMCWEYAMAWQKCMKNTLLLDAVIHCLSSISDIHIRLGLFSIIWATYIKQIFEISCRLVNKVGRLPKDPLCLQDISFDNTTLINFFRLTTLYLNDFQNCLKLSYSHNKKKVEFEKIWEESMPSLVEVTQHTKNVNDCILTLNYQVSCTLYYQCHFNIKLSKPLDVLFDIDYHYILEALTGNVVNREINMKPSEKLRNPRMKFLTKLIRASVETISCSDGHESYRTYDSGDCQMWIEKISILANLWSIDDEFIKRQQVIALYHVGYDTLAENTMNSIRNPVPVLAPILAITIQRLKRSLEHSNNQAEWIVSIPPQLYKRLQNTVLDTSIPAHPSLSTTILVLQKLLFQLNKKSPDPAHLQDIKLIELIIENCEIIIRRKL
Summary
Description
Probable regulatory subunit of a GTPase activating protein that has specificity for Rab3 subfamily. Rab3 proteins are involved in regulated exocytosis of neurotransmitters and hormones. Rab3 GTPase-activating complex specifically converts active Rab3-GTP to the inactive form Rab3-GDP (By similarity).
Catalytic Activity
ATP + (deoxyribonucleotide)(n)-3'-hydroxyl + 5'-phospho-(deoxyribonucleotide)(m) = (deoxyribonucleotide)(n+m) + AMP + diphosphate.
Subunit
The Rab3 GTPase-activating complex is a heterodimer composed of CG31935 and Rab3-GAP.
Similarity
Belongs to the ATP-dependent DNA ligase family.
Belongs to the Rab3-GAP regulatory subunit family.
Belongs to the Rab3-GAP regulatory subunit family.
Keywords
Complete proteome
Cytoplasm
GTPase activation
Phosphoprotein
Reference proteome
Feature
chain Rab3 GTPase-activating protein regulatory subunit
Uniprot
A0A2A4IZ58
A0A3S2TBU2
A0A212F3K2
A0A194PNF3
A0A194R3H1
A0A0L7LIW1
+ More
A0A2H1V5C5 A0A2A4J0Q1 A0A1Y1NCX5 A0A1Y1NCP5 A0A1W4XAX2 U4TTB4 D6X347 N6T5J7 A0A2J7R920 B0W297 A0A1L8DVY2 A0A182GWH7 A0A0T6AWP6 Q16V75 A0A1S4FMZ6 A0A0P6K0T9 A0A1B0FY92 A0A3L8DQ81 A0A0J7NWA9 A0A2A3E5F6 A0A1B6DPT0 A0A182R4X1 A0A182LRR3 A0A2P8Z6F9 E9IA54 A0A182ILN3 A0A1B0EVF1 A0A154PGB7 A0A0L7RC30 A0A182L9H5 A0A182WS99 A0A182K6M7 A0A182HL49 A0A182VWH5 K7IQJ3 A0A182U7Z4 Q380I7 A0A182PGW3 A0A232FEK1 A0A182UWM1 A0A195FVF7 A0A158NW53 A0A182NQE9 W4VRU7 A0A151J7H5 A0A182Q7S3 F4X586 A0A182YAG6 A0A0C9R1B1 A0A182FRH0 A0A151WVF2 W8C8C3 A0A0A1WZV5 A0A034VHQ1 E2BXV6 A0A1L8DVB9 B4N799 A0A1B6CRQ7 Q9VKB9 A0A3B0JKV0 A0A3B0JGC7 A0A1W4VB05 B3N3X8 B4P1Y0 B4Q3P5 B4IE58 A0A2J7R922 A0A0J9TLU2 B4KHZ4 A0A087ZMU0 A0A1B0G281 Q29NK1 A0A1B0BV61 B3MVI4 A0A182G8G5 B4GQJ8 A0A1A9XFP4 W8BCN5 A0A1I8MK59 A0A1A9UPY4 E2AI71 A0A2R5L7F0 A0A1I8Q0Z2 A0A091H3H3 F6V9A8 B4JBL0 F7GE41 A0A091PRW0 A0A1A7Y699 G3RS87 A0A2R9C7L3 A0A2K6PHJ7
A0A2H1V5C5 A0A2A4J0Q1 A0A1Y1NCX5 A0A1Y1NCP5 A0A1W4XAX2 U4TTB4 D6X347 N6T5J7 A0A2J7R920 B0W297 A0A1L8DVY2 A0A182GWH7 A0A0T6AWP6 Q16V75 A0A1S4FMZ6 A0A0P6K0T9 A0A1B0FY92 A0A3L8DQ81 A0A0J7NWA9 A0A2A3E5F6 A0A1B6DPT0 A0A182R4X1 A0A182LRR3 A0A2P8Z6F9 E9IA54 A0A182ILN3 A0A1B0EVF1 A0A154PGB7 A0A0L7RC30 A0A182L9H5 A0A182WS99 A0A182K6M7 A0A182HL49 A0A182VWH5 K7IQJ3 A0A182U7Z4 Q380I7 A0A182PGW3 A0A232FEK1 A0A182UWM1 A0A195FVF7 A0A158NW53 A0A182NQE9 W4VRU7 A0A151J7H5 A0A182Q7S3 F4X586 A0A182YAG6 A0A0C9R1B1 A0A182FRH0 A0A151WVF2 W8C8C3 A0A0A1WZV5 A0A034VHQ1 E2BXV6 A0A1L8DVB9 B4N799 A0A1B6CRQ7 Q9VKB9 A0A3B0JKV0 A0A3B0JGC7 A0A1W4VB05 B3N3X8 B4P1Y0 B4Q3P5 B4IE58 A0A2J7R922 A0A0J9TLU2 B4KHZ4 A0A087ZMU0 A0A1B0G281 Q29NK1 A0A1B0BV61 B3MVI4 A0A182G8G5 B4GQJ8 A0A1A9XFP4 W8BCN5 A0A1I8MK59 A0A1A9UPY4 E2AI71 A0A2R5L7F0 A0A1I8Q0Z2 A0A091H3H3 F6V9A8 B4JBL0 F7GE41 A0A091PRW0 A0A1A7Y699 G3RS87 A0A2R9C7L3 A0A2K6PHJ7
EC Number
6.5.1.1
Pubmed
22118469
26354079
26227816
28004739
23537049
18362917
+ More
19820115 26483478 17510324 26999592 30249741 29403074 21282665 20966253 20075255 12364791 28648823 21347285 21719571 25244985 24495485 25830018 25348373 20798317 17994087 10731132 12537572 10731138 10798391 18327897 17550304 22936249 15632085 25315136 17495919 18464734 22398555 22722832 25362486
19820115 26483478 17510324 26999592 30249741 29403074 21282665 20966253 20075255 12364791 28648823 21347285 21719571 25244985 24495485 25830018 25348373 20798317 17994087 10731132 12537572 10731138 10798391 18327897 17550304 22936249 15632085 25315136 17495919 18464734 22398555 22722832 25362486
EMBL
NWSH01004441
PCG65105.1
RSAL01000597
RVE41337.1
AGBW02010564
OWR48311.1
+ More
KQ459603 KPI92650.1 KQ460845 KPJ12069.1 JTDY01000955 KOB75289.1 ODYU01000758 SOQ36028.1 PCG65104.1 GEZM01006579 JAV95711.1 GEZM01006578 JAV95712.1 KB631008 KB632204 ERL83997.1 ERL90088.1 KQ971372 EFA10326.1 APGK01043446 KB741014 ENN75474.1 NEVH01006721 PNF37325.1 DS231826 EDS28712.1 GFDF01003505 JAV10579.1 JXUM01093231 JXUM01093232 KQ564043 KXJ72873.1 LJIG01022683 KRT79269.1 CH477602 EAT38427.1 GDUN01000773 JAN95146.1 AJVK01000819 AJVK01000820 QOIP01000005 RLU22590.1 LBMM01001228 KMQ96685.1 KZ288363 PBC26930.1 GEDC01009620 JAS27678.1 AXCM01000956 PYGN01000174 PSN52085.1 GL761987 EFZ22535.1 AJWK01020170 KQ434899 KZC10899.1 KQ414617 KOC68340.1 APCN01000881 AAAB01008987 EAL38577.4 NNAY01000360 OXU28898.1 KQ981215 KYN44625.1 ADTU01027812 GANO01000437 JAB59434.1 KQ979679 KYN19865.1 AXCN02001530 GL888695 EGI58384.1 GBYB01001709 GBYB01001717 GBYB01006672 JAG71476.1 JAG71484.1 JAG76439.1 KQ982706 KYQ51818.1 GAMC01007556 JAB98999.1 GBXI01013560 GBXI01010111 JAD00732.1 JAD04181.1 GAKP01017869 JAC41083.1 GL451323 EFN79470.1 GFDF01003714 JAV10370.1 CH964182 EDW80240.1 GEDC01021251 JAS16047.1 AE014134 BT016089 AF145682 OUUW01000006 SPP81443.1 SPP81444.1 CH954177 EDV58830.1 CM000157 EDW88151.1 CM000361 EDX04770.1 CH480831 EDW45885.1 PNF37324.1 CM002910 KMY89870.1 CH933807 EDW11276.1 CCAG010001456 CH379059 EAL34218.2 JXJN01021143 CH902624 EDV33249.1 JXUM01047743 JXUM01047744 JXUM01047745 JXUM01047746 JXUM01047747 KQ561531 KXJ78251.1 CH479187 EDW39870.1 GAMC01007555 JAB99000.1 GL439655 EFN66870.1 GGLE01001320 MBY05446.1 KL448134 KFO80827.1 CH916368 EDW04033.1 KK663996 KFQ10018.1 HADX01003740 SBP25972.1 CABD030008962 CABD030008963 CABD030008964 CABD030008965 CABD030008966 AJFE02020675 AJFE02020676 AJFE02020677
KQ459603 KPI92650.1 KQ460845 KPJ12069.1 JTDY01000955 KOB75289.1 ODYU01000758 SOQ36028.1 PCG65104.1 GEZM01006579 JAV95711.1 GEZM01006578 JAV95712.1 KB631008 KB632204 ERL83997.1 ERL90088.1 KQ971372 EFA10326.1 APGK01043446 KB741014 ENN75474.1 NEVH01006721 PNF37325.1 DS231826 EDS28712.1 GFDF01003505 JAV10579.1 JXUM01093231 JXUM01093232 KQ564043 KXJ72873.1 LJIG01022683 KRT79269.1 CH477602 EAT38427.1 GDUN01000773 JAN95146.1 AJVK01000819 AJVK01000820 QOIP01000005 RLU22590.1 LBMM01001228 KMQ96685.1 KZ288363 PBC26930.1 GEDC01009620 JAS27678.1 AXCM01000956 PYGN01000174 PSN52085.1 GL761987 EFZ22535.1 AJWK01020170 KQ434899 KZC10899.1 KQ414617 KOC68340.1 APCN01000881 AAAB01008987 EAL38577.4 NNAY01000360 OXU28898.1 KQ981215 KYN44625.1 ADTU01027812 GANO01000437 JAB59434.1 KQ979679 KYN19865.1 AXCN02001530 GL888695 EGI58384.1 GBYB01001709 GBYB01001717 GBYB01006672 JAG71476.1 JAG71484.1 JAG76439.1 KQ982706 KYQ51818.1 GAMC01007556 JAB98999.1 GBXI01013560 GBXI01010111 JAD00732.1 JAD04181.1 GAKP01017869 JAC41083.1 GL451323 EFN79470.1 GFDF01003714 JAV10370.1 CH964182 EDW80240.1 GEDC01021251 JAS16047.1 AE014134 BT016089 AF145682 OUUW01000006 SPP81443.1 SPP81444.1 CH954177 EDV58830.1 CM000157 EDW88151.1 CM000361 EDX04770.1 CH480831 EDW45885.1 PNF37324.1 CM002910 KMY89870.1 CH933807 EDW11276.1 CCAG010001456 CH379059 EAL34218.2 JXJN01021143 CH902624 EDV33249.1 JXUM01047743 JXUM01047744 JXUM01047745 JXUM01047746 JXUM01047747 KQ561531 KXJ78251.1 CH479187 EDW39870.1 GAMC01007555 JAB99000.1 GL439655 EFN66870.1 GGLE01001320 MBY05446.1 KL448134 KFO80827.1 CH916368 EDW04033.1 KK663996 KFQ10018.1 HADX01003740 SBP25972.1 CABD030008962 CABD030008963 CABD030008964 CABD030008965 CABD030008966 AJFE02020675 AJFE02020676 AJFE02020677
Proteomes
UP000218220
UP000283053
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000192223 UP000030742 UP000007266 UP000019118 UP000235965 UP000002320 UP000069940 UP000249989 UP000008820 UP000092462 UP000279307 UP000036403 UP000242457 UP000075900 UP000075883 UP000245037 UP000075880 UP000092461 UP000076502 UP000053825 UP000075882 UP000076407 UP000075881 UP000075840 UP000075920 UP000002358 UP000075902 UP000007062 UP000075885 UP000215335 UP000075903 UP000078541 UP000005205 UP000075884 UP000078492 UP000075886 UP000007755 UP000076408 UP000069272 UP000075809 UP000008237 UP000007798 UP000000803 UP000268350 UP000192221 UP000008711 UP000002282 UP000000304 UP000001292 UP000009192 UP000005203 UP000092444 UP000001819 UP000092460 UP000007801 UP000008744 UP000092443 UP000095301 UP000078200 UP000000311 UP000095300 UP000053760 UP000002280 UP000001070 UP000002279 UP000001519 UP000240080 UP000233200
UP000192223 UP000030742 UP000007266 UP000019118 UP000235965 UP000002320 UP000069940 UP000249989 UP000008820 UP000092462 UP000279307 UP000036403 UP000242457 UP000075900 UP000075883 UP000245037 UP000075880 UP000092461 UP000076502 UP000053825 UP000075882 UP000076407 UP000075881 UP000075840 UP000075920 UP000002358 UP000075902 UP000007062 UP000075885 UP000215335 UP000075903 UP000078541 UP000005205 UP000075884 UP000078492 UP000075886 UP000007755 UP000076408 UP000069272 UP000075809 UP000008237 UP000007798 UP000000803 UP000268350 UP000192221 UP000008711 UP000002282 UP000000304 UP000001292 UP000009192 UP000005203 UP000092444 UP000001819 UP000092460 UP000007801 UP000008744 UP000092443 UP000095301 UP000078200 UP000000311 UP000095300 UP000053760 UP000002280 UP000001070 UP000002279 UP000001519 UP000240080 UP000233200
PRIDE
Pfam
Interpro
IPR029257
RAB3GAP2_C
+ More
IPR026059 Rab3-gap_reg
IPR032839 RAB3GAP_N
IPR011047 Quinoprotein_ADH-like_supfam
IPR036322 WD40_repeat_dom_sf
IPR000977 DNA_ligase_ATP-dep
IPR012308 DNA_ligase_ATP-dep_N
IPR012309 DNA_ligase_ATP-dep_C
IPR036599 DNA_ligase_N_sf
IPR012310 DNA_ligase_ATP-dep_cent
IPR012340 NA-bd_OB-fold
IPR016059 DNA_ligase_ATP-dep_CS
IPR026059 Rab3-gap_reg
IPR032839 RAB3GAP_N
IPR011047 Quinoprotein_ADH-like_supfam
IPR036322 WD40_repeat_dom_sf
IPR000977 DNA_ligase_ATP-dep
IPR012308 DNA_ligase_ATP-dep_N
IPR012309 DNA_ligase_ATP-dep_C
IPR036599 DNA_ligase_N_sf
IPR012310 DNA_ligase_ATP-dep_cent
IPR012340 NA-bd_OB-fold
IPR016059 DNA_ligase_ATP-dep_CS
Gene 3D
ProteinModelPortal
A0A2A4IZ58
A0A3S2TBU2
A0A212F3K2
A0A194PNF3
A0A194R3H1
A0A0L7LIW1
+ More
A0A2H1V5C5 A0A2A4J0Q1 A0A1Y1NCX5 A0A1Y1NCP5 A0A1W4XAX2 U4TTB4 D6X347 N6T5J7 A0A2J7R920 B0W297 A0A1L8DVY2 A0A182GWH7 A0A0T6AWP6 Q16V75 A0A1S4FMZ6 A0A0P6K0T9 A0A1B0FY92 A0A3L8DQ81 A0A0J7NWA9 A0A2A3E5F6 A0A1B6DPT0 A0A182R4X1 A0A182LRR3 A0A2P8Z6F9 E9IA54 A0A182ILN3 A0A1B0EVF1 A0A154PGB7 A0A0L7RC30 A0A182L9H5 A0A182WS99 A0A182K6M7 A0A182HL49 A0A182VWH5 K7IQJ3 A0A182U7Z4 Q380I7 A0A182PGW3 A0A232FEK1 A0A182UWM1 A0A195FVF7 A0A158NW53 A0A182NQE9 W4VRU7 A0A151J7H5 A0A182Q7S3 F4X586 A0A182YAG6 A0A0C9R1B1 A0A182FRH0 A0A151WVF2 W8C8C3 A0A0A1WZV5 A0A034VHQ1 E2BXV6 A0A1L8DVB9 B4N799 A0A1B6CRQ7 Q9VKB9 A0A3B0JKV0 A0A3B0JGC7 A0A1W4VB05 B3N3X8 B4P1Y0 B4Q3P5 B4IE58 A0A2J7R922 A0A0J9TLU2 B4KHZ4 A0A087ZMU0 A0A1B0G281 Q29NK1 A0A1B0BV61 B3MVI4 A0A182G8G5 B4GQJ8 A0A1A9XFP4 W8BCN5 A0A1I8MK59 A0A1A9UPY4 E2AI71 A0A2R5L7F0 A0A1I8Q0Z2 A0A091H3H3 F6V9A8 B4JBL0 F7GE41 A0A091PRW0 A0A1A7Y699 G3RS87 A0A2R9C7L3 A0A2K6PHJ7
A0A2H1V5C5 A0A2A4J0Q1 A0A1Y1NCX5 A0A1Y1NCP5 A0A1W4XAX2 U4TTB4 D6X347 N6T5J7 A0A2J7R920 B0W297 A0A1L8DVY2 A0A182GWH7 A0A0T6AWP6 Q16V75 A0A1S4FMZ6 A0A0P6K0T9 A0A1B0FY92 A0A3L8DQ81 A0A0J7NWA9 A0A2A3E5F6 A0A1B6DPT0 A0A182R4X1 A0A182LRR3 A0A2P8Z6F9 E9IA54 A0A182ILN3 A0A1B0EVF1 A0A154PGB7 A0A0L7RC30 A0A182L9H5 A0A182WS99 A0A182K6M7 A0A182HL49 A0A182VWH5 K7IQJ3 A0A182U7Z4 Q380I7 A0A182PGW3 A0A232FEK1 A0A182UWM1 A0A195FVF7 A0A158NW53 A0A182NQE9 W4VRU7 A0A151J7H5 A0A182Q7S3 F4X586 A0A182YAG6 A0A0C9R1B1 A0A182FRH0 A0A151WVF2 W8C8C3 A0A0A1WZV5 A0A034VHQ1 E2BXV6 A0A1L8DVB9 B4N799 A0A1B6CRQ7 Q9VKB9 A0A3B0JKV0 A0A3B0JGC7 A0A1W4VB05 B3N3X8 B4P1Y0 B4Q3P5 B4IE58 A0A2J7R922 A0A0J9TLU2 B4KHZ4 A0A087ZMU0 A0A1B0G281 Q29NK1 A0A1B0BV61 B3MVI4 A0A182G8G5 B4GQJ8 A0A1A9XFP4 W8BCN5 A0A1I8MK59 A0A1A9UPY4 E2AI71 A0A2R5L7F0 A0A1I8Q0Z2 A0A091H3H3 F6V9A8 B4JBL0 F7GE41 A0A091PRW0 A0A1A7Y699 G3RS87 A0A2R9C7L3 A0A2K6PHJ7
Ontologies
GO
GO:0043087
GO:0017112
GO:0003910
GO:0005524
GO:0051103
GO:0006310
GO:0071897
GO:0006260
GO:0003677
GO:0008021
GO:0060025
GO:0046982
GO:0007298
GO:0007269
GO:0005096
GO:0030234
GO:0016192
GO:0097051
GO:2000786
GO:0005829
GO:0032991
GO:0005886
GO:1903061
GO:1903373
GO:0005515
GO:0006367
GO:0003975
GO:0006487
GO:0006488
GO:0007156
GO:0008237
GO:0043401
GO:0003707
GO:0051537
PANTHER
Topology
Subcellular location
Cytoplasm
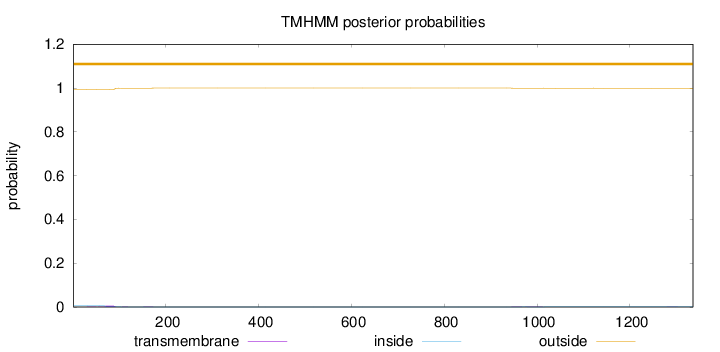
Length:
1336
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.191400000000001
Exp number, first 60 AAs:
0.01355
Total prob of N-in:
0.00623
outside
1 - 1336
Population Genetic Test Statistics
Pi
4.018947
Theta
7.935772
Tajima's D
-1.67836
CLR
1.309422
CSRT
0.0386480675966202
Interpretation
Uncertain
