Gene
KWMTBOMO07318
Annotation
glycoprotein_hormone_alpha_2_precursor_[Bombyx_mori]
Full name
Thyrostimulin alpha-2 subunit
Location in the cell
Extracellular Reliability : 2.214 PlasmaMembrane Reliability : 1.841
Sequence
CDS
ATGCTTCTTAGATTTATTGTTACCCTCATGTTTTTATGCCAAATTTTTGCCGCTGAATCGTGGAGAAAACCAGGCTGTCATAGAATTGGTCACACAAGAAATATAAGCATCCCAGATTGCGTAGAATTTAAAATAACAACGAATGCATGCCGCGGGTATTGTGAGTCCTGGTCCCTTCCCAGTATTATGCTTGGGTTTAAGAGACATCCCGTAACTTCATTAGGACAATGCTGTAACATAATGGAAGCTGAAGATGTACCTGTCAAAGTCCTGTGCTTGGATGGAGAAAGAAATTTAATATTTAAATCTGCGGTTTCATGCGCATGTTACCATTGCCAAAAAGAATGA
Protein
MLLRFIVTLMFLCQIFAAESWRKPGCHRIGHTRNISIPDCVEFKITTNACRGYCESWSLPSIMLGFKRHPVTSLGQCCNIMEAEDVPVKVLCLDGERNLIFKSAVSCACYHCQKE
Summary
Subunit
Heterodimer with GPHB5; non-covalently-linked.
Similarity
Belongs to the glycoprotein hormones subunit alpha family.
Keywords
Direct protein sequencing
Disulfide bond
Hormone
Secreted
Signal
Toxin
Feature
chain Thyrostimulin alpha-2 subunit
Uniprot
B3XYE1
A0A0L7LPR3
A0A194R3H7
A0A194PIV1
M4QC45
A0A0S1U1N8
+ More
A0A2A4J493 A0A385H8V5 A0A212F3K0 A0A2H1V2K4 A0A067QSH1 A0A2J7QXV2 C6SUP2 T1HSS2 W5J3I6 U3U7C9 A0A224XWZ9 A0A0H3YEI7 A0A182GQ02 A0A182PAU6 A0A2S2QRP7 A0A1Y9HEH9 A0A182JTH5 A0A2C9GS92 A0A182VEV2 A0A182TT19 A0A182L0T8 Q7PJ83 A0A182F505 A0A182Y3E8 B0X0Y3 A0A1E1G7S7 A0A182NU68 A0A182QNT0 D6X356 A0A182MUL9 A0A084VXY8 A0A2R7W268 C6SUP0 J9LHE2 A0A2U8JAF2 A0A1Q3FRB0 A0A1Q3FQ52 A0A1I8QBB4 A0A2S2NY71 A0A1Q3FQA2 A0A1I8M647 A0A0K8UIA1 W8CB56 A0A0L0C2Q8 A0A0P8XKI1 A0A1B0DQU1 A0A0A1X2W0 B4KFF3 B5DK43 A0A1W4UXI7 A0A2P8YSC7 A0A3B0JTX5 B4JQ28 B4MUL4 T1JQM9 Q58L88 A0A1S3CUF9 A0A0Q5VKA1 A0A0R1E961 A0A0P6BIF0 B2Z9X2 A0A1B0FQT8 A0A1A9VF46 A0A1A9W5J4 E9GLP7 A0A1D2NDK3 A0A1J1IT51 A0A182W6N4 A0A0P5QCG0 C6SUN9 A0A2T7NXA4 A0A0F7Z3J2 A0A1S6JQ29 A0A0P6IC84 A0A346CIW0 A0A346CJI7 A0A182J389 A0A087TXD7 A0A1Y3ANS8 C6SUP1 A0A1X9ZNE1 A0A0P5NNQ3 A0A2C9K1U4 A0A346GAT9 A0A336MA94 B4LQP5 A0A1X9ZND6 C6SUN8 O46182 K1QSB9 V3ZQ40 A0A1A9ZFC5
A0A2A4J493 A0A385H8V5 A0A212F3K0 A0A2H1V2K4 A0A067QSH1 A0A2J7QXV2 C6SUP2 T1HSS2 W5J3I6 U3U7C9 A0A224XWZ9 A0A0H3YEI7 A0A182GQ02 A0A182PAU6 A0A2S2QRP7 A0A1Y9HEH9 A0A182JTH5 A0A2C9GS92 A0A182VEV2 A0A182TT19 A0A182L0T8 Q7PJ83 A0A182F505 A0A182Y3E8 B0X0Y3 A0A1E1G7S7 A0A182NU68 A0A182QNT0 D6X356 A0A182MUL9 A0A084VXY8 A0A2R7W268 C6SUP0 J9LHE2 A0A2U8JAF2 A0A1Q3FRB0 A0A1Q3FQ52 A0A1I8QBB4 A0A2S2NY71 A0A1Q3FQA2 A0A1I8M647 A0A0K8UIA1 W8CB56 A0A0L0C2Q8 A0A0P8XKI1 A0A1B0DQU1 A0A0A1X2W0 B4KFF3 B5DK43 A0A1W4UXI7 A0A2P8YSC7 A0A3B0JTX5 B4JQ28 B4MUL4 T1JQM9 Q58L88 A0A1S3CUF9 A0A0Q5VKA1 A0A0R1E961 A0A0P6BIF0 B2Z9X2 A0A1B0FQT8 A0A1A9VF46 A0A1A9W5J4 E9GLP7 A0A1D2NDK3 A0A1J1IT51 A0A182W6N4 A0A0P5QCG0 C6SUN9 A0A2T7NXA4 A0A0F7Z3J2 A0A1S6JQ29 A0A0P6IC84 A0A346CIW0 A0A346CJI7 A0A182J389 A0A087TXD7 A0A1Y3ANS8 C6SUP1 A0A1X9ZNE1 A0A0P5NNQ3 A0A2C9K1U4 A0A346GAT9 A0A336MA94 B4LQP5 A0A1X9ZND6 C6SUN8 O46182 K1QSB9 V3ZQ40 A0A1A9ZFC5
Pubmed
19264871
26227816
26354079
30171635
22118469
24845553
+ More
20920257 23761445 23932938 26036749 26483478 20966253 12364791 14747013 17210077 25244985 18362917 19820115 24438588 20566863 27924858 25315136 24495485 26108605 17994087 25830018 15632085 29403074 10731132 12537568 12537572 12537573 12537574 15890769 16110336 17569856 17569867 26109357 26109356 17550304 21292972 27289101 24505301 26301480 28082215 30060147 28502716 15562597 9364772 22992520 23254933
20920257 23761445 23932938 26036749 26483478 20966253 12364791 14747013 17210077 25244985 18362917 19820115 24438588 20566863 27924858 25315136 24495485 26108605 17994087 25830018 15632085 29403074 10731132 12537568 12537572 12537573 12537574 15890769 16110336 17569856 17569867 26109357 26109356 17550304 21292972 27289101 24505301 26301480 28082215 30060147 28502716 15562597 9364772 22992520 23254933
EMBL
AB365128
BN001243
BAG55000.1
CAR94699.1
JTDY01000386
KOB77427.1
+ More
KQ460845 KPJ12074.1 KQ459603 KPI92644.1 KC340925 AGH25557.1 KT005971 RSAL01000022 ALM30322.1 RVE52390.1 NWSH01003090 PCG66967.1 MH028086 AXY04255.1 AGBW02010564 OWR48317.1 ODYU01000383 SOQ35077.1 KK853336 KDR08333.1 NEVH01009372 PNF33409.1 BN001241 CAR94697.1 ACPB03004813 ADMH02002110 ETN58907.1 AB817258 BAO00955.1 GFTR01002008 JAW14418.1 KP895540 AKN21239.1 JXUM01079391 KQ563124 KXJ74437.1 GGMS01011198 MBY80401.1 APCN01000797 AAAB01008964 EAA43859.4 BN001242 DS232248 CAR94698.1 EDS38375.1 LC146505 BAV78809.1 AXCN02000773 KQ971372 EFA10706.1 AXCM01004580 ATLV01018225 KE525226 KFB42832.1 KK854234 PTY13518.1 BN001239 DS235354 CAR94695.1 EEB15080.1 ABLF02034426 ABLF02034428 MH210978 AWK57521.1 GFDL01005013 JAV30032.1 GFDL01005381 JAV29664.1 GGMR01009403 MBY22022.1 GFDL01005254 JAV29791.1 GDHF01025882 JAI26432.1 GAMC01000759 JAC05797.1 JRES01000973 KNC26638.1 CH902628 KPU75323.1 AJVK01008750 GBXI01008628 JAD05664.1 CH933807 EDW12053.1 CH379061 EDY70669.1 PYGN01000390 PSN47146.1 OUUW01000010 SPP85555.1 CH916372 EDV99008.1 CH963857 EDW76209.1 CAEY01000437 AY940435 CP007081 AAX38184.1 EDP27970.2 CH954179 KQS61761.1 CH898078 KRK05705.1 GDIP01014090 GDIQ01079018 LRGB01000626 JAM89625.1 JAN15719.1 KZS17589.1 EU620226 ACC99601.1 CCAG010000954 GL732551 EFX79541.1 LJIJ01000077 ODN03327.1 CVRI01000058 CRL02756.1 GDIQ01137497 JAL14229.1 BN001238 CAR94694.2 PZQS01000008 PVD25804.1 GAIH01000105 KY287983 AQS80514.1 GDIQ01006694 JAN88043.1 MH360460 AXL95509.1 MH360687 AXL95736.1 KK117181 KFM69776.1 MUJZ01067344 OTF70089.1 BN001240 CAR94696.1 MF033101 ARS73219.1 GDIQ01139840 JAL11886.1 MH045224 AXN93492.1 UFQS01000558 UFQS01000675 UFQT01000558 UFQT01000675 SSX04925.1 SSX06070.1 SSX25287.1 CH940649 EDW63429.2 MF033099 ARS73217.1 FJ195453 BN001237 ACN32202.1 CAR94693.2 U73802 AAB92391.1 JH817298 EKC24491.1 KB201891 ESO93503.1
KQ460845 KPJ12074.1 KQ459603 KPI92644.1 KC340925 AGH25557.1 KT005971 RSAL01000022 ALM30322.1 RVE52390.1 NWSH01003090 PCG66967.1 MH028086 AXY04255.1 AGBW02010564 OWR48317.1 ODYU01000383 SOQ35077.1 KK853336 KDR08333.1 NEVH01009372 PNF33409.1 BN001241 CAR94697.1 ACPB03004813 ADMH02002110 ETN58907.1 AB817258 BAO00955.1 GFTR01002008 JAW14418.1 KP895540 AKN21239.1 JXUM01079391 KQ563124 KXJ74437.1 GGMS01011198 MBY80401.1 APCN01000797 AAAB01008964 EAA43859.4 BN001242 DS232248 CAR94698.1 EDS38375.1 LC146505 BAV78809.1 AXCN02000773 KQ971372 EFA10706.1 AXCM01004580 ATLV01018225 KE525226 KFB42832.1 KK854234 PTY13518.1 BN001239 DS235354 CAR94695.1 EEB15080.1 ABLF02034426 ABLF02034428 MH210978 AWK57521.1 GFDL01005013 JAV30032.1 GFDL01005381 JAV29664.1 GGMR01009403 MBY22022.1 GFDL01005254 JAV29791.1 GDHF01025882 JAI26432.1 GAMC01000759 JAC05797.1 JRES01000973 KNC26638.1 CH902628 KPU75323.1 AJVK01008750 GBXI01008628 JAD05664.1 CH933807 EDW12053.1 CH379061 EDY70669.1 PYGN01000390 PSN47146.1 OUUW01000010 SPP85555.1 CH916372 EDV99008.1 CH963857 EDW76209.1 CAEY01000437 AY940435 CP007081 AAX38184.1 EDP27970.2 CH954179 KQS61761.1 CH898078 KRK05705.1 GDIP01014090 GDIQ01079018 LRGB01000626 JAM89625.1 JAN15719.1 KZS17589.1 EU620226 ACC99601.1 CCAG010000954 GL732551 EFX79541.1 LJIJ01000077 ODN03327.1 CVRI01000058 CRL02756.1 GDIQ01137497 JAL14229.1 BN001238 CAR94694.2 PZQS01000008 PVD25804.1 GAIH01000105 KY287983 AQS80514.1 GDIQ01006694 JAN88043.1 MH360460 AXL95509.1 MH360687 AXL95736.1 KK117181 KFM69776.1 MUJZ01067344 OTF70089.1 BN001240 CAR94696.1 MF033101 ARS73219.1 GDIQ01139840 JAL11886.1 MH045224 AXN93492.1 UFQS01000558 UFQS01000675 UFQT01000558 UFQT01000675 SSX04925.1 SSX06070.1 SSX25287.1 CH940649 EDW63429.2 MF033099 ARS73217.1 FJ195453 BN001237 ACN32202.1 CAR94693.2 U73802 AAB92391.1 JH817298 EKC24491.1 KB201891 ESO93503.1
Proteomes
UP000037510
UP000053240
UP000053268
UP000283053
UP000218220
UP000007151
+ More
UP000027135 UP000235965 UP000015103 UP000000673 UP000069940 UP000249989 UP000075885 UP000075900 UP000075881 UP000075840 UP000075903 UP000075902 UP000075882 UP000007062 UP000069272 UP000076408 UP000002320 UP000075884 UP000075886 UP000007266 UP000075883 UP000030765 UP000009046 UP000007819 UP000095300 UP000095301 UP000037069 UP000007801 UP000092462 UP000009192 UP000001819 UP000192221 UP000245037 UP000268350 UP000001070 UP000007798 UP000015104 UP000000803 UP000079169 UP000008711 UP000002282 UP000076858 UP000092444 UP000078200 UP000091820 UP000000305 UP000094527 UP000183832 UP000075920 UP000245119 UP000075880 UP000054359 UP000076420 UP000008792 UP000005408 UP000030746 UP000092445
UP000027135 UP000235965 UP000015103 UP000000673 UP000069940 UP000249989 UP000075885 UP000075900 UP000075881 UP000075840 UP000075903 UP000075902 UP000075882 UP000007062 UP000069272 UP000076408 UP000002320 UP000075884 UP000075886 UP000007266 UP000075883 UP000030765 UP000009046 UP000007819 UP000095300 UP000095301 UP000037069 UP000007801 UP000092462 UP000009192 UP000001819 UP000192221 UP000245037 UP000268350 UP000001070 UP000007798 UP000015104 UP000000803 UP000079169 UP000008711 UP000002282 UP000076858 UP000092444 UP000078200 UP000091820 UP000000305 UP000094527 UP000183832 UP000075920 UP000245119 UP000075880 UP000054359 UP000076420 UP000008792 UP000005408 UP000030746 UP000092445
Pfam
Interpro
Gene 3D
ProteinModelPortal
B3XYE1
A0A0L7LPR3
A0A194R3H7
A0A194PIV1
M4QC45
A0A0S1U1N8
+ More
A0A2A4J493 A0A385H8V5 A0A212F3K0 A0A2H1V2K4 A0A067QSH1 A0A2J7QXV2 C6SUP2 T1HSS2 W5J3I6 U3U7C9 A0A224XWZ9 A0A0H3YEI7 A0A182GQ02 A0A182PAU6 A0A2S2QRP7 A0A1Y9HEH9 A0A182JTH5 A0A2C9GS92 A0A182VEV2 A0A182TT19 A0A182L0T8 Q7PJ83 A0A182F505 A0A182Y3E8 B0X0Y3 A0A1E1G7S7 A0A182NU68 A0A182QNT0 D6X356 A0A182MUL9 A0A084VXY8 A0A2R7W268 C6SUP0 J9LHE2 A0A2U8JAF2 A0A1Q3FRB0 A0A1Q3FQ52 A0A1I8QBB4 A0A2S2NY71 A0A1Q3FQA2 A0A1I8M647 A0A0K8UIA1 W8CB56 A0A0L0C2Q8 A0A0P8XKI1 A0A1B0DQU1 A0A0A1X2W0 B4KFF3 B5DK43 A0A1W4UXI7 A0A2P8YSC7 A0A3B0JTX5 B4JQ28 B4MUL4 T1JQM9 Q58L88 A0A1S3CUF9 A0A0Q5VKA1 A0A0R1E961 A0A0P6BIF0 B2Z9X2 A0A1B0FQT8 A0A1A9VF46 A0A1A9W5J4 E9GLP7 A0A1D2NDK3 A0A1J1IT51 A0A182W6N4 A0A0P5QCG0 C6SUN9 A0A2T7NXA4 A0A0F7Z3J2 A0A1S6JQ29 A0A0P6IC84 A0A346CIW0 A0A346CJI7 A0A182J389 A0A087TXD7 A0A1Y3ANS8 C6SUP1 A0A1X9ZNE1 A0A0P5NNQ3 A0A2C9K1U4 A0A346GAT9 A0A336MA94 B4LQP5 A0A1X9ZND6 C6SUN8 O46182 K1QSB9 V3ZQ40 A0A1A9ZFC5
A0A2A4J493 A0A385H8V5 A0A212F3K0 A0A2H1V2K4 A0A067QSH1 A0A2J7QXV2 C6SUP2 T1HSS2 W5J3I6 U3U7C9 A0A224XWZ9 A0A0H3YEI7 A0A182GQ02 A0A182PAU6 A0A2S2QRP7 A0A1Y9HEH9 A0A182JTH5 A0A2C9GS92 A0A182VEV2 A0A182TT19 A0A182L0T8 Q7PJ83 A0A182F505 A0A182Y3E8 B0X0Y3 A0A1E1G7S7 A0A182NU68 A0A182QNT0 D6X356 A0A182MUL9 A0A084VXY8 A0A2R7W268 C6SUP0 J9LHE2 A0A2U8JAF2 A0A1Q3FRB0 A0A1Q3FQ52 A0A1I8QBB4 A0A2S2NY71 A0A1Q3FQA2 A0A1I8M647 A0A0K8UIA1 W8CB56 A0A0L0C2Q8 A0A0P8XKI1 A0A1B0DQU1 A0A0A1X2W0 B4KFF3 B5DK43 A0A1W4UXI7 A0A2P8YSC7 A0A3B0JTX5 B4JQ28 B4MUL4 T1JQM9 Q58L88 A0A1S3CUF9 A0A0Q5VKA1 A0A0R1E961 A0A0P6BIF0 B2Z9X2 A0A1B0FQT8 A0A1A9VF46 A0A1A9W5J4 E9GLP7 A0A1D2NDK3 A0A1J1IT51 A0A182W6N4 A0A0P5QCG0 C6SUN9 A0A2T7NXA4 A0A0F7Z3J2 A0A1S6JQ29 A0A0P6IC84 A0A346CIW0 A0A346CJI7 A0A182J389 A0A087TXD7 A0A1Y3ANS8 C6SUP1 A0A1X9ZNE1 A0A0P5NNQ3 A0A2C9K1U4 A0A346GAT9 A0A336MA94 B4LQP5 A0A1X9ZND6 C6SUN8 O46182 K1QSB9 V3ZQ40 A0A1A9ZFC5
Ontologies
GO
PANTHER
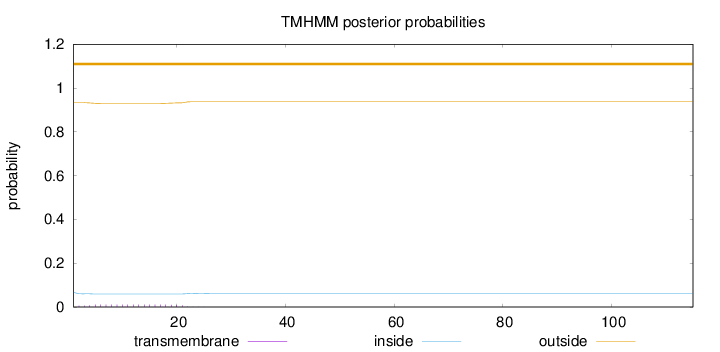
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 20,
Likelihood: 0.998057
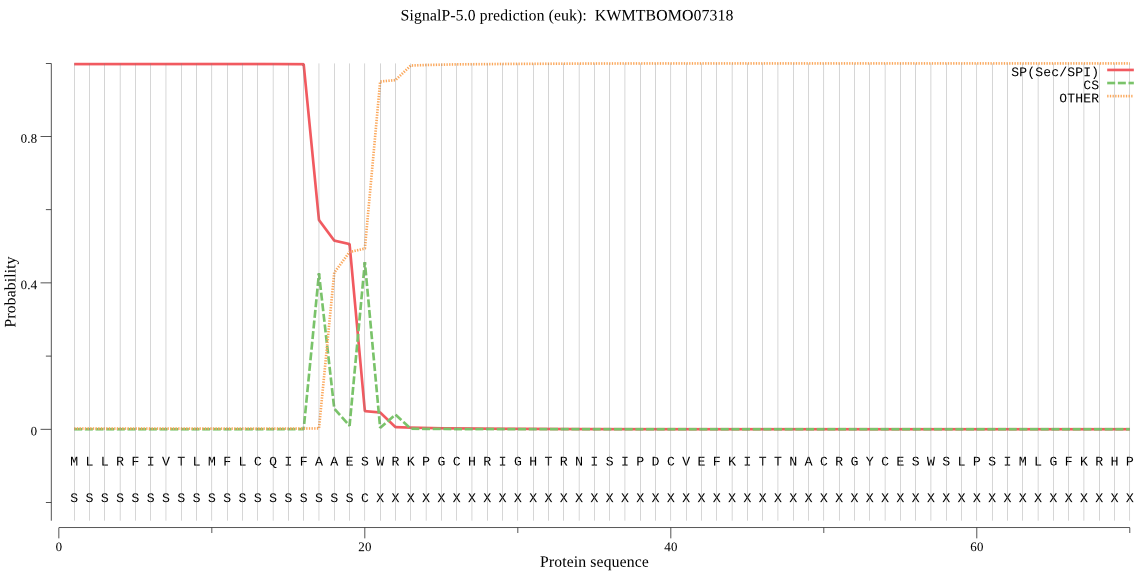
Length:
115
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.19954
Exp number, first 60 AAs:
0.19949
Total prob of N-in:
0.06665
outside
1 - 115
Population Genetic Test Statistics
Pi
6.317983
Theta
15.246379
Tajima's D
-1.0525
CLR
0.570526
CSRT
0.127743612819359
Interpretation
Uncertain
