Gene
KWMTBOMO07317
Pre Gene Modal
BGIBMGA010625
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_vacuolar_protein_sorting-associated_protein_54_[Bombyx_mori]
Full name
Vacuolar protein sorting-associated protein 54
Alternative Name
Protein scattered
Location in the cell
Nuclear Reliability : 2.953
Sequence
CDS
ATGGAAGATGTTAATGGGAGTGATAATGAAGAGATTTGGTCAGCTGAAGAATTGACATCTATTGAAGACAAATTGAAAAAAATGATTGTTTCATTGTCAGATTACTGCAATGAAAGATGTGCTAACCTAATTGTAACTAAAACAGACAAAGATTATGTGTGTTCTGATATGTCACAGTTATCTCTCCTATCAAATTTGATTAAAGAATTTTGCACCAAATGTGAGGATGTAACCGGATATTGTAGTAATGCCATGAATTTGGCTCTAAGGAGCTTTGCTATGAAGTATATACAGACACTTCACACTGAACGCAGATCTCAATTAACAGCTTCTCTAAATGCTGAACGATGGAAAACTGCAGATGTTCCATATGAATTGCAAAGTGCAATCACAAAAATATGTGAAATTCAAGAAATACCTCCTAATTTACTGTATGAAAGTGGGAAACCAGACGGAAAATATCTTTTTATAAACAAAGAAAGCTATGCTCTGGTAGGGACAGTTCAACTACTTATAAAAATTTTATTGGAATACTGTGATGCAACCAGACAATCTCCCATTATAGTTCAGTATTTAGTTCATTGTATGTTAGAATTATTAAGATTGTTTAATTCTCGGTGCTGCCAACTTGTTCTCGGTGCAGGGGCAATACAAAGTGCTGGACTTAAAACTATTTCTACATCCAATTTAGCTCTAGTATCTCGATCTTTACAGGTTATATTATGGTTTTTACCTATGATAAAACAGCTGTTAGAGAAAATACATTCTTCTGAGGTATCATTGAGCGGTTTTGTTGCTATTGAGAGTGATATTGCTAGCCATAAACAGGAAATAGAAAATAAGATTTGCTTAATTGTAAGTAATATGTTATGTTCTCAATTAACAGGGTGGGATGCAAAGCCCCCAGTTCCTTCTCAAACATTCCGCAATATTTCAAAACATTTAGTGAAGTTACATGAAGCATTAATTGATATTTTGCCACCTGAACAAATAAAGTTTATATACAAACGAGTACATGATAATTTTAAAGATAAATTAAGAGAACAACTAATTAAAATGAATATAGTTGCTAATGGAAGCCCCCAACATGGTGTAGTGACTTCTGAATTGACATTTTACTTACAAACTCTAAAGACACTCAGAGTTATAAACGATAATGATGCAGAAGACAATATACTATATGACGTTTGGTTAAATTAA
Protein
MEDVNGSDNEEIWSAEELTSIEDKLKKMIVSLSDYCNERCANLIVTKTDKDYVCSDMSQLSLLSNLIKEFCTKCEDVTGYCSNAMNLALRSFAMKYIQTLHTERRSQLTASLNAERWKTADVPYELQSAITKICEIQEIPPNLLYESGKPDGKYLFINKESYALVGTVQLLIKILLEYCDATRQSPIIVQYLVHCMLELLRLFNSRCCQLVLGAGAIQSAGLKTISTSNLALVSRSLQVILWFLPMIKQLLEKIHSSEVSLSGFVAIESDIASHKQEIENKICLIVSNMLCSQLTGWDAKPPVPSQTFRNISKHLVKLHEALIDILPPEQIKFIYKRVHDNFKDKLREQLIKMNIVANGSPQHGVVTSELTFYLQTLKTLRVINDNDAEDNILYDVWLN
Summary
Description
May be involved in retrograde transport from early and late endosomes to late Golgi (By similarity). Required during spermatogenesis for sperm individualization.
Similarity
Belongs to the VPS54 family.
Keywords
Coiled coil
Complete proteome
Developmental protein
Differentiation
Golgi apparatus
Phosphoprotein
Protein transport
Reference proteome
Spermatogenesis
Transport
Feature
chain Vacuolar protein sorting-associated protein 54
Uniprot
A0A0L7L7V6
A0A2A4J5L8
A0A2H1V2S6
A0A3S2TLN5
A0A212F3K1
A0A194R2Q7
+ More
A0A194PNE8 A0A1L8DVG9 B0WC20 A0A182H6U5 A0A182GWR3 A0A1Q3EX00 A0A0P6J3Y7 A0A1S4EYY6 A0A1J1I403 A0A182ISM8 A0A1B0CJL3 A0A1I8NBJ3 A0A182NTX3 E2C760 Q7Q4D8 A0A0L0C8E9 A0A034V9C9 U5EVN8 D6X241 A0A1I8PN64 A0A0T6B5V4 A0A0A1WT42 W8BBJ1 U4UE23 A0A0K8V6E5 A0A0K8WEU1 A0A1B0D2T3 A0A1W4WXB3 A0A0C9RDR0 A0A1Y1LRW8 A0A336LPB0 A0A1W4VHX0 A0A3B0JUV5 B4GKS6 Q29PH7 A0A0L7RH22 B4M9R9 A0A195FIM1 B3MUU3 A0A0J7L806 A0A151WTU9 A0A195DM28 B3N7Z8 A0A195BTB0 B4N1D6 E2AY30 A0A0J9R030 B4NY36 A0A1A9W025 B4HYX5 Q9VLC0 B4JPV9 A0A026WC72 A0A158P2E0 F4WUR4 A0A2J7QXU0 A0A1B0A4Z7 K7IWH9 E9IGC2 B4KIQ3 A0A0M5J3Z8 A0A195D384 A0A310S5Y0 A0A1B0FFB5 A0A1A9V9L8 A0A088A9H6 N6U2S3 A0A1A9XGV2 A0A1B0BJH1 A0A2A3EGK3 A0A0M9A7P7 A0A067QV92 R7UWE0 K1Q8F6 A0A087UES5 T1J950 A0A2D4LVJ8 A0A1B6DGB4 A0A210PHC0 A0A2D4PKF3 G3HDQ0 E9GYI9 A0A2D4HCZ4 A0A2D4HD00 A0A0N8A5N3 A0A2D4PKE5 S4RLY6 A0A0P6I3K3 A0A164TR75 A0A2D4LUI5 A0A2D4LUZ1 A0A1W7RBQ9
A0A194PNE8 A0A1L8DVG9 B0WC20 A0A182H6U5 A0A182GWR3 A0A1Q3EX00 A0A0P6J3Y7 A0A1S4EYY6 A0A1J1I403 A0A182ISM8 A0A1B0CJL3 A0A1I8NBJ3 A0A182NTX3 E2C760 Q7Q4D8 A0A0L0C8E9 A0A034V9C9 U5EVN8 D6X241 A0A1I8PN64 A0A0T6B5V4 A0A0A1WT42 W8BBJ1 U4UE23 A0A0K8V6E5 A0A0K8WEU1 A0A1B0D2T3 A0A1W4WXB3 A0A0C9RDR0 A0A1Y1LRW8 A0A336LPB0 A0A1W4VHX0 A0A3B0JUV5 B4GKS6 Q29PH7 A0A0L7RH22 B4M9R9 A0A195FIM1 B3MUU3 A0A0J7L806 A0A151WTU9 A0A195DM28 B3N7Z8 A0A195BTB0 B4N1D6 E2AY30 A0A0J9R030 B4NY36 A0A1A9W025 B4HYX5 Q9VLC0 B4JPV9 A0A026WC72 A0A158P2E0 F4WUR4 A0A2J7QXU0 A0A1B0A4Z7 K7IWH9 E9IGC2 B4KIQ3 A0A0M5J3Z8 A0A195D384 A0A310S5Y0 A0A1B0FFB5 A0A1A9V9L8 A0A088A9H6 N6U2S3 A0A1A9XGV2 A0A1B0BJH1 A0A2A3EGK3 A0A0M9A7P7 A0A067QV92 R7UWE0 K1Q8F6 A0A087UES5 T1J950 A0A2D4LVJ8 A0A1B6DGB4 A0A210PHC0 A0A2D4PKF3 G3HDQ0 E9GYI9 A0A2D4HCZ4 A0A2D4HD00 A0A0N8A5N3 A0A2D4PKE5 S4RLY6 A0A0P6I3K3 A0A164TR75 A0A2D4LUI5 A0A2D4LUZ1 A0A1W7RBQ9
Pubmed
26227816
22118469
26354079
26483478
26999592
25315136
+ More
20798317 12364791 26108605 25348373 18362917 19820115 25830018 24495485 23537049 28004739 17994087 15632085 22936249 17550304 10731132 12537572 12537569 9550716 18327897 24508170 30249741 21347285 21719571 20075255 21282665 24845553 23254933 22992520 28812685 21804562 21292972 26358130
20798317 12364791 26108605 25348373 18362917 19820115 25830018 24495485 23537049 28004739 17994087 15632085 22936249 17550304 10731132 12537572 12537569 9550716 18327897 24508170 30249741 21347285 21719571 20075255 21282665 24845553 23254933 22992520 28812685 21804562 21292972 26358130
EMBL
JTDY01002364
KOB71573.1
NWSH01003090
PCG66968.1
ODYU01000383
SOQ35076.1
+ More
RSAL01000063 RVE49484.1 AGBW02010564 OWR48316.1 KQ460845 KPJ12073.1 KQ459603 KPI92645.1 GFDF01003653 JAV10431.1 DS231884 EDS43080.1 JXUM01114915 KQ565831 KXJ70621.1 JXUM01093988 JXUM01093989 JXUM01093990 JXUM01093991 JXUM01093992 JXUM01093993 JXUM01093994 KQ564095 KXJ72804.1 GFDL01015213 JAV19832.1 GDUN01001103 JAN94816.1 CVRI01000037 CRK93598.1 AJWK01014705 AJWK01014706 GL453340 EFN76173.1 AAAB01008964 EAA12219.3 JRES01000864 KNC27679.1 GAKP01019823 JAC39129.1 GANO01001780 JAB58091.1 KQ971371 EFA09922.2 LJIG01009604 KRT82748.1 GBXI01012694 GBXI01003674 JAD01598.1 JAD10618.1 GAMC01012162 JAB94393.1 KB632100 ERL88826.1 GDHF01026324 GDHF01018016 JAI25990.1 JAI34298.1 GDHF01002697 JAI49617.1 AJVK01002795 AJVK01002796 AJVK01002797 AJVK01002798 AJVK01002799 AJVK01002800 GBYB01007242 GBYB01011237 JAG77009.1 JAG81004.1 GEZM01048806 JAV76393.1 UFQT01000093 SSX19625.1 OUUW01000010 SPP85885.1 CH479184 EDW37242.1 CH379058 EAL34316.1 KQ414594 KOC70138.1 CH940654 EDW57945.1 KQ981523 KYN40248.1 CH902624 EDV33008.1 LBMM01000329 KMR01092.1 KQ982753 KYQ51197.1 KQ980734 KYN13945.1 CH954177 EDV58359.1 KQ976408 KYM91198.1 CH963920 EDW78073.1 GL443762 EFN61626.1 CM002910 KMY89259.1 CM000157 EDW88638.1 CH480818 EDW52255.1 AE014134 AY061309 CH916372 EDV98939.1 KK107323 QOIP01000002 EZA52629.1 RLU25806.1 ADTU01007165 GL888375 EGI62061.1 NEVH01009372 PNF33403.1 GL762997 EFZ20380.1 CH933807 EDW12409.1 CP012525 ALC44468.1 KQ976885 KYN07357.1 KQ769058 OAD53014.1 CCAG010014241 APGK01041782 KB740997 ENN75870.1 JXJN01015395 KZ288254 PBC30838.1 KQ435730 KOX77559.1 KK853336 KDR08332.1 AMQN01005993 KB297336 ELU10634.1 JH817970 EKC25155.1 KK119496 KFM75864.1 JH431970 IACM01046221 LAB24713.1 GEDC01012565 JAS24733.1 NEDP02076698 OWF35898.1 IACN01072218 LAB58504.1 JH000301 EGW00356.1 GL732575 EFX75401.1 IACK01012036 LAA69830.1 IACK01012034 LAA69823.1 GDIP01180073 JAJ43329.1 IACN01072217 LAB58502.1 GDIQ01143161 GDIQ01011899 JAN82838.1 LRGB01001763 KZS10677.1 IACM01046222 LAB24715.1 IACM01046224 LAB24718.1 GDAY02002889 JAV48578.1
RSAL01000063 RVE49484.1 AGBW02010564 OWR48316.1 KQ460845 KPJ12073.1 KQ459603 KPI92645.1 GFDF01003653 JAV10431.1 DS231884 EDS43080.1 JXUM01114915 KQ565831 KXJ70621.1 JXUM01093988 JXUM01093989 JXUM01093990 JXUM01093991 JXUM01093992 JXUM01093993 JXUM01093994 KQ564095 KXJ72804.1 GFDL01015213 JAV19832.1 GDUN01001103 JAN94816.1 CVRI01000037 CRK93598.1 AJWK01014705 AJWK01014706 GL453340 EFN76173.1 AAAB01008964 EAA12219.3 JRES01000864 KNC27679.1 GAKP01019823 JAC39129.1 GANO01001780 JAB58091.1 KQ971371 EFA09922.2 LJIG01009604 KRT82748.1 GBXI01012694 GBXI01003674 JAD01598.1 JAD10618.1 GAMC01012162 JAB94393.1 KB632100 ERL88826.1 GDHF01026324 GDHF01018016 JAI25990.1 JAI34298.1 GDHF01002697 JAI49617.1 AJVK01002795 AJVK01002796 AJVK01002797 AJVK01002798 AJVK01002799 AJVK01002800 GBYB01007242 GBYB01011237 JAG77009.1 JAG81004.1 GEZM01048806 JAV76393.1 UFQT01000093 SSX19625.1 OUUW01000010 SPP85885.1 CH479184 EDW37242.1 CH379058 EAL34316.1 KQ414594 KOC70138.1 CH940654 EDW57945.1 KQ981523 KYN40248.1 CH902624 EDV33008.1 LBMM01000329 KMR01092.1 KQ982753 KYQ51197.1 KQ980734 KYN13945.1 CH954177 EDV58359.1 KQ976408 KYM91198.1 CH963920 EDW78073.1 GL443762 EFN61626.1 CM002910 KMY89259.1 CM000157 EDW88638.1 CH480818 EDW52255.1 AE014134 AY061309 CH916372 EDV98939.1 KK107323 QOIP01000002 EZA52629.1 RLU25806.1 ADTU01007165 GL888375 EGI62061.1 NEVH01009372 PNF33403.1 GL762997 EFZ20380.1 CH933807 EDW12409.1 CP012525 ALC44468.1 KQ976885 KYN07357.1 KQ769058 OAD53014.1 CCAG010014241 APGK01041782 KB740997 ENN75870.1 JXJN01015395 KZ288254 PBC30838.1 KQ435730 KOX77559.1 KK853336 KDR08332.1 AMQN01005993 KB297336 ELU10634.1 JH817970 EKC25155.1 KK119496 KFM75864.1 JH431970 IACM01046221 LAB24713.1 GEDC01012565 JAS24733.1 NEDP02076698 OWF35898.1 IACN01072218 LAB58504.1 JH000301 EGW00356.1 GL732575 EFX75401.1 IACK01012036 LAA69830.1 IACK01012034 LAA69823.1 GDIP01180073 JAJ43329.1 IACN01072217 LAB58502.1 GDIQ01143161 GDIQ01011899 JAN82838.1 LRGB01001763 KZS10677.1 IACM01046222 LAB24715.1 IACM01046224 LAB24718.1 GDAY02002889 JAV48578.1
Proteomes
UP000037510
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
+ More
UP000002320 UP000069940 UP000249989 UP000183832 UP000075880 UP000092461 UP000095301 UP000075884 UP000008237 UP000007062 UP000037069 UP000007266 UP000095300 UP000030742 UP000092462 UP000192223 UP000192221 UP000268350 UP000008744 UP000001819 UP000053825 UP000008792 UP000078541 UP000007801 UP000036403 UP000075809 UP000078492 UP000008711 UP000078540 UP000007798 UP000000311 UP000002282 UP000091820 UP000001292 UP000000803 UP000001070 UP000053097 UP000279307 UP000005205 UP000007755 UP000235965 UP000092445 UP000002358 UP000009192 UP000092553 UP000078542 UP000092444 UP000078200 UP000005203 UP000019118 UP000092443 UP000092460 UP000242457 UP000053105 UP000027135 UP000014760 UP000005408 UP000054359 UP000242188 UP000001075 UP000000305 UP000245300 UP000076858
UP000002320 UP000069940 UP000249989 UP000183832 UP000075880 UP000092461 UP000095301 UP000075884 UP000008237 UP000007062 UP000037069 UP000007266 UP000095300 UP000030742 UP000092462 UP000192223 UP000192221 UP000268350 UP000008744 UP000001819 UP000053825 UP000008792 UP000078541 UP000007801 UP000036403 UP000075809 UP000078492 UP000008711 UP000078540 UP000007798 UP000000311 UP000002282 UP000091820 UP000001292 UP000000803 UP000001070 UP000053097 UP000279307 UP000005205 UP000007755 UP000235965 UP000092445 UP000002358 UP000009192 UP000092553 UP000078542 UP000092444 UP000078200 UP000005203 UP000019118 UP000092443 UP000092460 UP000242457 UP000053105 UP000027135 UP000014760 UP000005408 UP000054359 UP000242188 UP000001075 UP000000305 UP000245300 UP000076858
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A0L7L7V6
A0A2A4J5L8
A0A2H1V2S6
A0A3S2TLN5
A0A212F3K1
A0A194R2Q7
+ More
A0A194PNE8 A0A1L8DVG9 B0WC20 A0A182H6U5 A0A182GWR3 A0A1Q3EX00 A0A0P6J3Y7 A0A1S4EYY6 A0A1J1I403 A0A182ISM8 A0A1B0CJL3 A0A1I8NBJ3 A0A182NTX3 E2C760 Q7Q4D8 A0A0L0C8E9 A0A034V9C9 U5EVN8 D6X241 A0A1I8PN64 A0A0T6B5V4 A0A0A1WT42 W8BBJ1 U4UE23 A0A0K8V6E5 A0A0K8WEU1 A0A1B0D2T3 A0A1W4WXB3 A0A0C9RDR0 A0A1Y1LRW8 A0A336LPB0 A0A1W4VHX0 A0A3B0JUV5 B4GKS6 Q29PH7 A0A0L7RH22 B4M9R9 A0A195FIM1 B3MUU3 A0A0J7L806 A0A151WTU9 A0A195DM28 B3N7Z8 A0A195BTB0 B4N1D6 E2AY30 A0A0J9R030 B4NY36 A0A1A9W025 B4HYX5 Q9VLC0 B4JPV9 A0A026WC72 A0A158P2E0 F4WUR4 A0A2J7QXU0 A0A1B0A4Z7 K7IWH9 E9IGC2 B4KIQ3 A0A0M5J3Z8 A0A195D384 A0A310S5Y0 A0A1B0FFB5 A0A1A9V9L8 A0A088A9H6 N6U2S3 A0A1A9XGV2 A0A1B0BJH1 A0A2A3EGK3 A0A0M9A7P7 A0A067QV92 R7UWE0 K1Q8F6 A0A087UES5 T1J950 A0A2D4LVJ8 A0A1B6DGB4 A0A210PHC0 A0A2D4PKF3 G3HDQ0 E9GYI9 A0A2D4HCZ4 A0A2D4HD00 A0A0N8A5N3 A0A2D4PKE5 S4RLY6 A0A0P6I3K3 A0A164TR75 A0A2D4LUI5 A0A2D4LUZ1 A0A1W7RBQ9
A0A194PNE8 A0A1L8DVG9 B0WC20 A0A182H6U5 A0A182GWR3 A0A1Q3EX00 A0A0P6J3Y7 A0A1S4EYY6 A0A1J1I403 A0A182ISM8 A0A1B0CJL3 A0A1I8NBJ3 A0A182NTX3 E2C760 Q7Q4D8 A0A0L0C8E9 A0A034V9C9 U5EVN8 D6X241 A0A1I8PN64 A0A0T6B5V4 A0A0A1WT42 W8BBJ1 U4UE23 A0A0K8V6E5 A0A0K8WEU1 A0A1B0D2T3 A0A1W4WXB3 A0A0C9RDR0 A0A1Y1LRW8 A0A336LPB0 A0A1W4VHX0 A0A3B0JUV5 B4GKS6 Q29PH7 A0A0L7RH22 B4M9R9 A0A195FIM1 B3MUU3 A0A0J7L806 A0A151WTU9 A0A195DM28 B3N7Z8 A0A195BTB0 B4N1D6 E2AY30 A0A0J9R030 B4NY36 A0A1A9W025 B4HYX5 Q9VLC0 B4JPV9 A0A026WC72 A0A158P2E0 F4WUR4 A0A2J7QXU0 A0A1B0A4Z7 K7IWH9 E9IGC2 B4KIQ3 A0A0M5J3Z8 A0A195D384 A0A310S5Y0 A0A1B0FFB5 A0A1A9V9L8 A0A088A9H6 N6U2S3 A0A1A9XGV2 A0A1B0BJH1 A0A2A3EGK3 A0A0M9A7P7 A0A067QV92 R7UWE0 K1Q8F6 A0A087UES5 T1J950 A0A2D4LVJ8 A0A1B6DGB4 A0A210PHC0 A0A2D4PKF3 G3HDQ0 E9GYI9 A0A2D4HCZ4 A0A2D4HD00 A0A0N8A5N3 A0A2D4PKE5 S4RLY6 A0A0P6I3K3 A0A164TR75 A0A2D4LUI5 A0A2D4LUZ1 A0A1W7RBQ9
PDB
3N1E
E-value=4.38474e-21,
Score=250
Ontologies
GO
PANTHER
Topology
Subcellular location
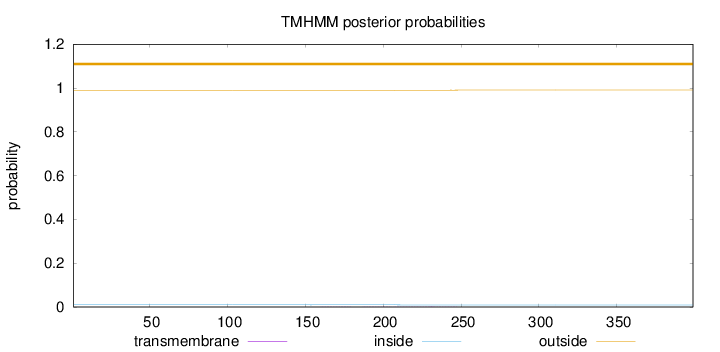
Length:
399
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04479
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01000
outside
1 - 399
Population Genetic Test Statistics
Pi
1.925696
Theta
4.635344
Tajima's D
-1.541327
CLR
1.085305
CSRT
0.0499975001249938
Interpretation
Uncertain
