Gene
KWMTBOMO07316
Pre Gene Modal
BGIBMGA010664
Annotation
PREDICTED:_UDP-N-acetylglucosamine--dolichyl-phosphate_N-acetylglucosaminephosphotransferase_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.993
Sequence
CDS
ATGTTATCATTGATATTATTGATGATTCTTTCATTGATAGCTTTTTTAATCACTGATGAACTTATTCATAAGTTAAAGAATTTATTCATAAACGCCGGCTTATATGGGATCGATTTATGCAAGACTGATAAAAATAAAATACCAGAGGCCTTAGGTGTTGTATCAGGATGTGTCTTCCTAATAACATGTTTTTTATTTATTCCCATAGCGTTTGGAAATGGATTAATGGATACACAACATTTTCCTCACAATGAATTTGCAGAATTTCTGGCAGCATTGTTATCAATTTGCTGCATGCTACTTCTTGGATTTGCTGATGATGTGTTAGATCTTAGATGGAGGTATAAACTTGTGCTTCCAACAGTGGCATCATTGCCATTACTTGTGGTGTACTATGTCAACTTTAATTCAACAACATTCATAATACCTATACCATTTAGATCGTGGTTCGGAGTTTCTGTGAATATTGGCTTTTTGTATTATATCTATATGGGTATGCTTGCTGTCTTCTGTACAAATGCAATCAATATCTTAGCTGGAATTAATGGTATAGAAGTAGGACAATCATTTGTCATTGCACTTTCCATTGTGGTATTTGATATTATAGAACTTCAAGGAGACCAATACAAGGCCCATACATTTTCTCTGCATATAATGATACCTTATTTGGCAACAACACTAGGACTGTTAAAACATAATTGGTATCCATCAAGAGTTTTTGTTGGTGATACGTTTTGCTATGTGTCAGGGATGACCTTTGCAGTTGTAGGCATTCTAAGTCATTTCAATATCCCTTAG
Protein
MLSLILLMILSLIAFLITDELIHKLKNLFINAGLYGIDLCKTDKNKIPEALGVVSGCVFLITCFLFIPIAFGNGLMDTQHFPHNEFAEFLAALLSICCMLLLGFADDVLDLRWRYKLVLPTVASLPLLVVYYVNFNSTTFIIPIPFRSWFGVSVNIGFLYYIYMGMLAVFCTNAINILAGINGIEVGQSFVIALSIVVFDIIELQGDQYKAHTFSLHIMIPYLATTLGLLKHNWYPSRVFVGDTFCYVSGMTFAVVGILSHFNIP
Summary
Uniprot
A0A2H1V2K3
A0A2A4J509
A0A212F3K7
S4PEM0
A0A194R306
A0A194PGX7
+ More
M4HZR9 A0A0L7L8G6 A0A067RTU0 A0A154PQ17 A0A2J7Q566 D2A2H3 A0A1Y1JUD0 A0A0L7R5Y8 A0A232FEN7 A0A0M9A5R3 K7IZS7 A0A151IGV7 A0A2S2QMH6 A0A158NBI3 A0A1B6KJR6 A0A1B6F1B3 A0A1B6CB88 A0A1B6HS77 A0A2A3EM29 A0A195DR66 A0A1W4XJQ2 E2C6L8 A0A3L8DT55 A0A195B9P2 A0A195FCV1 A0A2H8TZX5 A0A023F4S9 A0A224Z399 A0A0K8RMN2 T1I8K5 A0A023GE01 A0A2S2P3Z6 G3MP66 A0A131Z6D9 A0A146KU90 A0A0A9XJT6 A0A069DTB4 J9K1P6 A0A0K8RD07 A0A2P8YVN4 U5EL52 E0W3S4 A0A0K8SKU3 A0A023FKR2 A0A1L8E1E8 A0A293N303 V4BB73 A0A2R7W5Z2 A0A087SX13 A0A2R5LLP5 A0A1L8E1U2 T1JP24 B4LT07 T2M638 A0A1I8QBK9 A0A139WJ57 A0A2P6L9C0 A0A1I8MIH5 A0A2C9JEI9 A0A2I0LK30 A0A0M4ECT1 T1P8D3 M7BCW3 A0A1V4J8B0 B4IEG0 A0A084W887 A0A218UPY8 K7G9H9 A0A0Q3M7T4 B0W1F7 H9GMJ6 A0A182UNG9 A0A182WU17 Q7Q406 A0A182HUN0 B3N394 A0A182PPJ7 B4P2M3 A0A182F4W2 Q9VK30 A0A1W7R8H4 A0A182QE79 R7U1S9 A0A210PR81 A0A3L8S164 A0A182IR77 A0A1U7SH59 A0A1Q3F2Q0 A0A0K8TNH0 A0A182Y092 A0A0J9R1N5 A0A3M0IYU7 A0A3Q7SF59
M4HZR9 A0A0L7L8G6 A0A067RTU0 A0A154PQ17 A0A2J7Q566 D2A2H3 A0A1Y1JUD0 A0A0L7R5Y8 A0A232FEN7 A0A0M9A5R3 K7IZS7 A0A151IGV7 A0A2S2QMH6 A0A158NBI3 A0A1B6KJR6 A0A1B6F1B3 A0A1B6CB88 A0A1B6HS77 A0A2A3EM29 A0A195DR66 A0A1W4XJQ2 E2C6L8 A0A3L8DT55 A0A195B9P2 A0A195FCV1 A0A2H8TZX5 A0A023F4S9 A0A224Z399 A0A0K8RMN2 T1I8K5 A0A023GE01 A0A2S2P3Z6 G3MP66 A0A131Z6D9 A0A146KU90 A0A0A9XJT6 A0A069DTB4 J9K1P6 A0A0K8RD07 A0A2P8YVN4 U5EL52 E0W3S4 A0A0K8SKU3 A0A023FKR2 A0A1L8E1E8 A0A293N303 V4BB73 A0A2R7W5Z2 A0A087SX13 A0A2R5LLP5 A0A1L8E1U2 T1JP24 B4LT07 T2M638 A0A1I8QBK9 A0A139WJ57 A0A2P6L9C0 A0A1I8MIH5 A0A2C9JEI9 A0A2I0LK30 A0A0M4ECT1 T1P8D3 M7BCW3 A0A1V4J8B0 B4IEG0 A0A084W887 A0A218UPY8 K7G9H9 A0A0Q3M7T4 B0W1F7 H9GMJ6 A0A182UNG9 A0A182WU17 Q7Q406 A0A182HUN0 B3N394 A0A182PPJ7 B4P2M3 A0A182F4W2 Q9VK30 A0A1W7R8H4 A0A182QE79 R7U1S9 A0A210PR81 A0A3L8S164 A0A182IR77 A0A1U7SH59 A0A1Q3F2Q0 A0A0K8TNH0 A0A182Y092 A0A0J9R1N5 A0A3M0IYU7 A0A3Q7SF59
Pubmed
22118469
23622113
26354079
26227816
24845553
18362917
+ More
19820115 28004739 28648823 20075255 21347285 20798317 30249741 25474469 28797301 22216098 26830274 26823975 25401762 26334808 29403074 20566863 23254933 17994087 24065732 25315136 15562597 23371554 23624526 24438588 17381049 21881562 12364791 14747013 17210077 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28812685 30282656 26369729 25244985 22936249
19820115 28004739 28648823 20075255 21347285 20798317 30249741 25474469 28797301 22216098 26830274 26823975 25401762 26334808 29403074 20566863 23254933 17994087 24065732 25315136 15562597 23371554 23624526 24438588 17381049 21881562 12364791 14747013 17210077 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28812685 30282656 26369729 25244985 22936249
EMBL
ODYU01000383
SOQ35075.1
NWSH01003090
PCG66969.1
AGBW02010564
OWR48315.1
+ More
GAIX01007005 JAA85555.1 KQ460845 KPJ12072.1 KQ459603 KPI92646.1 JQ768053 AFV61654.1 JTDY01002364 KOB71576.1 KK852424 KDR24230.1 KQ435030 KZC14005.1 NEVH01018372 PNF23716.1 KQ971338 EFA02198.2 GEZM01100401 GEZM01100398 GEZM01100397 JAV52904.1 KQ414648 KOC66258.1 NNAY01000319 OXU29254.1 KQ435728 KOX77790.1 KQ977661 KYN00696.1 GGMS01009754 MBY78957.1 ADTU01011111 ADTU01011112 GEBQ01028583 JAT11394.1 GECZ01026106 JAS43663.1 GEDC01026557 JAS10741.1 GECU01030178 GECU01011605 JAS77528.1 JAS96101.1 KZ288219 PBC32256.1 KQ980581 KYN15405.1 GL453160 EFN76420.1 QOIP01000004 RLU23516.1 KQ976540 KYM81248.1 KQ981673 KYN38238.1 GFXV01006893 MBW18698.1 GBBI01002704 JAC16008.1 GFPF01013180 MAA24326.1 GADI01001685 JAA72123.1 ACPB03020207 GBBM01003331 JAC32087.1 GGMR01011453 MBY24072.1 JO843667 AEO35284.1 GEDV01002202 JAP86355.1 GDHC01018761 JAP99867.1 GBHO01022622 GBHO01022621 JAG20982.1 JAG20983.1 GBGD01001699 JAC87190.1 ABLF02034137 ABLF02034139 GADI01004847 JAA68961.1 PYGN01000334 PSN48313.1 GANO01001570 JAB58301.1 DS235882 EEB20281.1 GBRD01012095 JAG53729.1 GBBK01002743 JAC21739.1 GFDF01001600 JAV12484.1 GFWV01022716 MAA47443.1 KB199651 ESP04791.1 KK854341 PTY14848.1 KK112341 KFM57402.1 GGLE01006318 MBY10444.1 GFDF01001599 JAV12485.1 JH430694 CH940649 EDW63838.1 HAAD01001163 CDG67395.1 KYB27827.1 MWRG01000850 PRD35161.1 AKCR02000268 PKK17794.1 CP012523 ALC39485.1 KA644884 AFP59513.1 KB536242 EMP33405.1 LSYS01008642 OPJ68339.1 CH480831 EDW45987.1 ATLV01021389 KE525317 KFB46431.1 MUZQ01000188 OWK55716.1 AGCU01062366 AGCU01062367 LMAW01002639 KQK78733.1 DS231821 EDS45027.1 AAWZ02036114 AAAB01008964 EAA12410.3 APCN01002467 CH954177 EDV58734.1 CM000157 EDW88253.1 AE014134 AY051538 AAF53252.2 AAK92962.1 GEHC01000198 JAV47447.1 AXCN02000762 AMQN01011011 KB308639 ELT97140.1 NEDP02005551 OWF38952.1 QUSF01000089 RLV93731.1 GFDL01013236 JAV21809.1 GDAI01002128 JAI15475.1 CM002910 KMY90026.1 QRBI01000231 RMB91913.1
GAIX01007005 JAA85555.1 KQ460845 KPJ12072.1 KQ459603 KPI92646.1 JQ768053 AFV61654.1 JTDY01002364 KOB71576.1 KK852424 KDR24230.1 KQ435030 KZC14005.1 NEVH01018372 PNF23716.1 KQ971338 EFA02198.2 GEZM01100401 GEZM01100398 GEZM01100397 JAV52904.1 KQ414648 KOC66258.1 NNAY01000319 OXU29254.1 KQ435728 KOX77790.1 KQ977661 KYN00696.1 GGMS01009754 MBY78957.1 ADTU01011111 ADTU01011112 GEBQ01028583 JAT11394.1 GECZ01026106 JAS43663.1 GEDC01026557 JAS10741.1 GECU01030178 GECU01011605 JAS77528.1 JAS96101.1 KZ288219 PBC32256.1 KQ980581 KYN15405.1 GL453160 EFN76420.1 QOIP01000004 RLU23516.1 KQ976540 KYM81248.1 KQ981673 KYN38238.1 GFXV01006893 MBW18698.1 GBBI01002704 JAC16008.1 GFPF01013180 MAA24326.1 GADI01001685 JAA72123.1 ACPB03020207 GBBM01003331 JAC32087.1 GGMR01011453 MBY24072.1 JO843667 AEO35284.1 GEDV01002202 JAP86355.1 GDHC01018761 JAP99867.1 GBHO01022622 GBHO01022621 JAG20982.1 JAG20983.1 GBGD01001699 JAC87190.1 ABLF02034137 ABLF02034139 GADI01004847 JAA68961.1 PYGN01000334 PSN48313.1 GANO01001570 JAB58301.1 DS235882 EEB20281.1 GBRD01012095 JAG53729.1 GBBK01002743 JAC21739.1 GFDF01001600 JAV12484.1 GFWV01022716 MAA47443.1 KB199651 ESP04791.1 KK854341 PTY14848.1 KK112341 KFM57402.1 GGLE01006318 MBY10444.1 GFDF01001599 JAV12485.1 JH430694 CH940649 EDW63838.1 HAAD01001163 CDG67395.1 KYB27827.1 MWRG01000850 PRD35161.1 AKCR02000268 PKK17794.1 CP012523 ALC39485.1 KA644884 AFP59513.1 KB536242 EMP33405.1 LSYS01008642 OPJ68339.1 CH480831 EDW45987.1 ATLV01021389 KE525317 KFB46431.1 MUZQ01000188 OWK55716.1 AGCU01062366 AGCU01062367 LMAW01002639 KQK78733.1 DS231821 EDS45027.1 AAWZ02036114 AAAB01008964 EAA12410.3 APCN01002467 CH954177 EDV58734.1 CM000157 EDW88253.1 AE014134 AY051538 AAF53252.2 AAK92962.1 GEHC01000198 JAV47447.1 AXCN02000762 AMQN01011011 KB308639 ELT97140.1 NEDP02005551 OWF38952.1 QUSF01000089 RLV93731.1 GFDL01013236 JAV21809.1 GDAI01002128 JAI15475.1 CM002910 KMY90026.1 QRBI01000231 RMB91913.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
UP000027135
+ More
UP000076502 UP000235965 UP000007266 UP000053825 UP000215335 UP000053105 UP000002358 UP000078542 UP000005205 UP000242457 UP000078492 UP000192223 UP000008237 UP000279307 UP000078540 UP000078541 UP000015103 UP000007819 UP000245037 UP000009046 UP000030746 UP000054359 UP000008792 UP000095300 UP000095301 UP000076420 UP000053872 UP000092553 UP000031443 UP000190648 UP000001292 UP000030765 UP000197619 UP000007267 UP000051836 UP000002320 UP000001646 UP000075903 UP000076407 UP000007062 UP000075840 UP000008711 UP000075885 UP000002282 UP000069272 UP000000803 UP000075886 UP000014760 UP000242188 UP000276834 UP000075880 UP000189705 UP000076408 UP000269221 UP000286640
UP000076502 UP000235965 UP000007266 UP000053825 UP000215335 UP000053105 UP000002358 UP000078542 UP000005205 UP000242457 UP000078492 UP000192223 UP000008237 UP000279307 UP000078540 UP000078541 UP000015103 UP000007819 UP000245037 UP000009046 UP000030746 UP000054359 UP000008792 UP000095300 UP000095301 UP000076420 UP000053872 UP000092553 UP000031443 UP000190648 UP000001292 UP000030765 UP000197619 UP000007267 UP000051836 UP000002320 UP000001646 UP000075903 UP000076407 UP000007062 UP000075840 UP000008711 UP000075885 UP000002282 UP000069272 UP000000803 UP000075886 UP000014760 UP000242188 UP000276834 UP000075880 UP000189705 UP000076408 UP000269221 UP000286640
PRIDE
Pfam
PF00953 Glycos_transf_4
ProteinModelPortal
A0A2H1V2K3
A0A2A4J509
A0A212F3K7
S4PEM0
A0A194R306
A0A194PGX7
+ More
M4HZR9 A0A0L7L8G6 A0A067RTU0 A0A154PQ17 A0A2J7Q566 D2A2H3 A0A1Y1JUD0 A0A0L7R5Y8 A0A232FEN7 A0A0M9A5R3 K7IZS7 A0A151IGV7 A0A2S2QMH6 A0A158NBI3 A0A1B6KJR6 A0A1B6F1B3 A0A1B6CB88 A0A1B6HS77 A0A2A3EM29 A0A195DR66 A0A1W4XJQ2 E2C6L8 A0A3L8DT55 A0A195B9P2 A0A195FCV1 A0A2H8TZX5 A0A023F4S9 A0A224Z399 A0A0K8RMN2 T1I8K5 A0A023GE01 A0A2S2P3Z6 G3MP66 A0A131Z6D9 A0A146KU90 A0A0A9XJT6 A0A069DTB4 J9K1P6 A0A0K8RD07 A0A2P8YVN4 U5EL52 E0W3S4 A0A0K8SKU3 A0A023FKR2 A0A1L8E1E8 A0A293N303 V4BB73 A0A2R7W5Z2 A0A087SX13 A0A2R5LLP5 A0A1L8E1U2 T1JP24 B4LT07 T2M638 A0A1I8QBK9 A0A139WJ57 A0A2P6L9C0 A0A1I8MIH5 A0A2C9JEI9 A0A2I0LK30 A0A0M4ECT1 T1P8D3 M7BCW3 A0A1V4J8B0 B4IEG0 A0A084W887 A0A218UPY8 K7G9H9 A0A0Q3M7T4 B0W1F7 H9GMJ6 A0A182UNG9 A0A182WU17 Q7Q406 A0A182HUN0 B3N394 A0A182PPJ7 B4P2M3 A0A182F4W2 Q9VK30 A0A1W7R8H4 A0A182QE79 R7U1S9 A0A210PR81 A0A3L8S164 A0A182IR77 A0A1U7SH59 A0A1Q3F2Q0 A0A0K8TNH0 A0A182Y092 A0A0J9R1N5 A0A3M0IYU7 A0A3Q7SF59
M4HZR9 A0A0L7L8G6 A0A067RTU0 A0A154PQ17 A0A2J7Q566 D2A2H3 A0A1Y1JUD0 A0A0L7R5Y8 A0A232FEN7 A0A0M9A5R3 K7IZS7 A0A151IGV7 A0A2S2QMH6 A0A158NBI3 A0A1B6KJR6 A0A1B6F1B3 A0A1B6CB88 A0A1B6HS77 A0A2A3EM29 A0A195DR66 A0A1W4XJQ2 E2C6L8 A0A3L8DT55 A0A195B9P2 A0A195FCV1 A0A2H8TZX5 A0A023F4S9 A0A224Z399 A0A0K8RMN2 T1I8K5 A0A023GE01 A0A2S2P3Z6 G3MP66 A0A131Z6D9 A0A146KU90 A0A0A9XJT6 A0A069DTB4 J9K1P6 A0A0K8RD07 A0A2P8YVN4 U5EL52 E0W3S4 A0A0K8SKU3 A0A023FKR2 A0A1L8E1E8 A0A293N303 V4BB73 A0A2R7W5Z2 A0A087SX13 A0A2R5LLP5 A0A1L8E1U2 T1JP24 B4LT07 T2M638 A0A1I8QBK9 A0A139WJ57 A0A2P6L9C0 A0A1I8MIH5 A0A2C9JEI9 A0A2I0LK30 A0A0M4ECT1 T1P8D3 M7BCW3 A0A1V4J8B0 B4IEG0 A0A084W887 A0A218UPY8 K7G9H9 A0A0Q3M7T4 B0W1F7 H9GMJ6 A0A182UNG9 A0A182WU17 Q7Q406 A0A182HUN0 B3N394 A0A182PPJ7 B4P2M3 A0A182F4W2 Q9VK30 A0A1W7R8H4 A0A182QE79 R7U1S9 A0A210PR81 A0A3L8S164 A0A182IR77 A0A1U7SH59 A0A1Q3F2Q0 A0A0K8TNH0 A0A182Y092 A0A0J9R1N5 A0A3M0IYU7 A0A3Q7SF59
PDB
6FM9
E-value=7.34891e-74,
Score=703
Ontologies
PATHWAY
GO
PANTHER
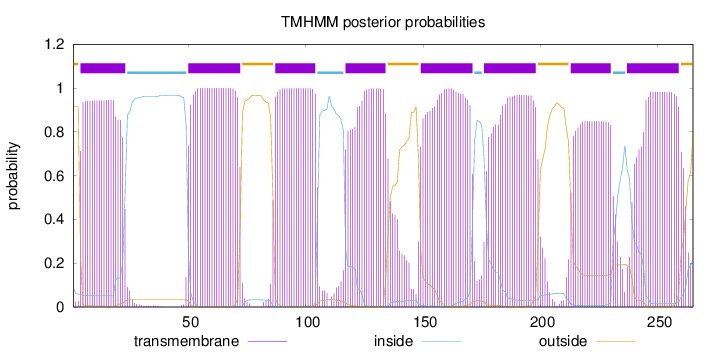
Topology
Length:
265
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
163.19873
Exp number, first 60 AAs:
28.80757
Total prob of N-in:
0.08511
POSSIBLE N-term signal
sequence
outside
1 - 3
TMhelix
4 - 23
inside
24 - 49
TMhelix
50 - 72
outside
73 - 86
TMhelix
87 - 104
inside
105 - 116
TMhelix
117 - 134
outside
135 - 148
TMhelix
149 - 171
inside
172 - 175
TMhelix
176 - 198
outside
199 - 212
TMhelix
213 - 230
inside
231 - 236
TMhelix
237 - 259
outside
260 - 265
Population Genetic Test Statistics
Pi
5.496761
Theta
13.498555
Tajima's D
-1.064459
CLR
0.020695
CSRT
0.126393680315984
Interpretation
Uncertain
