Gene
KWMTBOMO07305
Pre Gene Modal
BGIBMGA010659
Annotation
Noki_protein_[Operophtera_brumata]
Location in the cell
Extracellular Reliability : 1.234 Nuclear Reliability : 1.139
Sequence
CDS
ATGAGGTGGTTAATAGTGACAATCAGTGTGATGTTATCAAAAACAATACTGGCTGGGGTAGCGCCTCCCGGTTCGTGTCCTGCTGTTTGTGCGTGCAAATGGAAAGGAGGCAAACAAACCGTGGAATGCGTAGATAGAGCTCTCATCACGGTACCAGAACCGGTGGATCCTGCCACGCAAGTACTTGACCTTTCAGGAAATAATCTCCAAATATTACCACAGGAGGCTTTTGCGAGAACTGGACTGGTTAATTTACAAAGAGTATATCTTCGAAGTTGTAACATTGGTCAGATACATGACAGAGCATTCAAAGGGTTGACAAATTTAGTGGAGTTAGATTTGTCGCAGAATTTATTGACACAGATTCCATCAAACAGTTTCAAAGATGCACCTTTTTTACGAGACCTCACCCTGGCTCAAAATCCGATATTAAAAGTTCATTCTGATGCTTTGACAAATCTTGGAAGCGTTATTAAACTAGATTTATCCAGATGCGAAATTAGAGATGTAGCTGCAGATGCATTTAGAAATTTACGTTCAATGGAATCACTTAAGCTTGATCATAACAGACTTCGAGAATTTCCATTGAATTCTATTGAAAAAATTGAAAAATTGAGAGCTATTGATTTATCGGATAATCCTTGGATATGTGATTGTCGGCTTCGTGACCTGAAAATATGGTTAGCCAAAAACAAAATGCTTTCAACTCCTAGTTGTTCTGCACCAGCAAGATTGACGGGTCGACCATTTTCAGAGTTAGCTATTGAAGAGTTTGCGTGTAAGCCCGAAATATTACCTATAAGTCGGCATATTGAAGCAAATGTGGGGGAAAATACCACGATAACTTGTAGAACTGAAGCTATACCAAGTGCAGTAATAAACTGGTACTGGAATGGTAGATTACTTCAAAATGGTAGTCATTTTAACTCCCATCAAAAAATATACATTTTCGAAGAAGGCGATGCAAAAAAGAGGTCAATATTGATTCTCACGAACACACAGGAATCCGATTCCAGTGAATTTTATTGCGTTGCCGAAAACAAGGGTGGTAACGCGGAAGCTAACTTTACAGTACACGTTACGCAAATTGCAGTCGGAATGGCGTCACTTGGTAATGCTCAGATTGCGAGTTTGGGAGCGGTATTGTTTTTAATAATCATCGCTGTTTGTTTGGCAACTCTAGCTACATTTCTGCGGCTTCGTACGACCCCTGTTTGTGATACTAAAACTCCTAACACATTGGATCGCGTGGTATCAGGAAACGAAGTTCATCCGTCAAATAATGACCGTCCGCAAGTAGCTGTACTGGCTAATAGACAGGAATCATCGAATTACCGTGATTCTAAATGTAATCCTGTGATGAAACCGCCAAGAGTGAATGATATACCATATACGACTAACCACTACGAGGGAAGAGGAAGCGTTGTGACTGCGAACGGACCTGTTGTGGTTTCACCGACGGTGTCCGCTTCGATCGACCCTGATCTTATCAACGACACAAGGCCGGATAGTGCTAATAGGCCAGGGAGCGGCGAGTACGCACGCGAGGCTTCTGATTCTTTATATCCATCTGGATTGTGGGATCAAGTTAAATTGAATCAGACAAATAATTTGTCTCGGGCTATAAGTTCAGCTATACCCACATACTATAACGACAGAACACCGATAATCGAGAATGGAAGTGTGAACGGTTCTCAGGAAGAGTTGGGCTATATGAGTCGAACCTTCCCACGATCTCATGCGCTTTCTGCCACAAATACAGTGGTATCCACAGATGCCCCTTACCCACCCGACTATGGCCTGCCAGTAGGAGGCGCGCGCACTCTCCGTGTGTGGCAGCGTGCACCCCCGGTTCTGCCTCCAGTGTCAGCGTTAAAACGCGTTCTTACAATCACCAGGCCGTCTGCAGATGACGGCTTTCAAGATGGCTGTGCTACAGACGTTTAA
Protein
MRWLIVTISVMLSKTILAGVAPPGSCPAVCACKWKGGKQTVECVDRALITVPEPVDPATQVLDLSGNNLQILPQEAFARTGLVNLQRVYLRSCNIGQIHDRAFKGLTNLVELDLSQNLLTQIPSNSFKDAPFLRDLTLAQNPILKVHSDALTNLGSVIKLDLSRCEIRDVAADAFRNLRSMESLKLDHNRLREFPLNSIEKIEKLRAIDLSDNPWICDCRLRDLKIWLAKNKMLSTPSCSAPARLTGRPFSELAIEEFACKPEILPISRHIEANVGENTTITCRTEAIPSAVINWYWNGRLLQNGSHFNSHQKIYIFEEGDAKKRSILILTNTQESDSSEFYCVAENKGGNAEANFTVHVTQIAVGMASLGNAQIASLGAVLFLIIIAVCLATLATFLRLRTTPVCDTKTPNTLDRVVSGNEVHPSNNDRPQVAVLANRQESSNYRDSKCNPVMKPPRVNDIPYTTNHYEGRGSVVTANGPVVVSPTVSASIDPDLINDTRPDSANRPGSGEYAREASDSLYPSGLWDQVKLNQTNNLSRAISSAIPTYYNDRTPIIENGSVNGSQEELGYMSRTFPRSHALSATNTVVSTDAPYPPDYGLPVGGARTLRVWQRAPPVLPPVSALKRVLTITRPSADDGFQDGCATDV
Summary
Uniprot
H9JMA6
A0A2H1VV08
A0A0L7KT99
A0A2A4J6M8
A0A212EUM8
A0A194PH25
+ More
A0A194R895 A0A1Y1KPV8 A0A1B0GQT1 B0X983 D2A216 Q17LD1 A0A1S4EYJ9 A0A1B6D059 U4U6V4 A0A2J7RQF6 A0A2P8ZNT7 N6UDM9 A0A084WC07 A0A1J1HEI3 A0A182ITX6 N6URS7 T1HFN4 A0A182I527 A0A182U8F1 A0A182WYP4 A0A182TB69 A0A182R9K7 A0A182YCJ8 A0A182P6T4 A0A182NF07 A0A182WH30 A0A182MP10 A0A146LEI1 A0A1W4WR46 A0A0A9WHB9 A0A182KHJ8 A0A336LZT5 A0A1S4H0Z5 W5JD82 A0A067RBY4 A0A182F5D2 A0A182KUK4 A0A2R7WXQ2 N6TXI0 A0A182V957 Q7PVZ6 A0A0A1XT83 A0A0K8WG59 A0A1W4V6X6 Q8T3J2 B4MWL0 B3MNI3 Q2XY32 P91643 Q9VK54 Q2XY29 Q2XY28 Q2XY30 Q2XY31 B4IEC7 B4P2J5 L7XBB4 A0A0J9R1M2 B4HCX9 Q2XY27 B3N3C1 B4JA35 B4LST1 Q29L68 A0A3B0KAM7 A0A2A3EKJ6 B4Q498 B4KGL2 A0A087ZPS3 A0A0M9A3L1 A0A310S8P8 A0A1S3D0I3 A0A1I8MH62 A0A0L0BX47 E2BHT5 A0A1A9Z7U9 A0A1A9V871 A0A1B0BAG3 E2AXW2 A0A1I8P7S1 A0A1A9XEN7 A0A1A9WXL5 A0A195EDT3 A0A195EUQ4 A0A151IP40 A0A158NXN6 A0A0J7KRU9 A0A3L8E423 A0A026WJP7 A0A2H8TQI4 A0A2S2NE99 J9K4V7 A0A154P0C2
A0A194R895 A0A1Y1KPV8 A0A1B0GQT1 B0X983 D2A216 Q17LD1 A0A1S4EYJ9 A0A1B6D059 U4U6V4 A0A2J7RQF6 A0A2P8ZNT7 N6UDM9 A0A084WC07 A0A1J1HEI3 A0A182ITX6 N6URS7 T1HFN4 A0A182I527 A0A182U8F1 A0A182WYP4 A0A182TB69 A0A182R9K7 A0A182YCJ8 A0A182P6T4 A0A182NF07 A0A182WH30 A0A182MP10 A0A146LEI1 A0A1W4WR46 A0A0A9WHB9 A0A182KHJ8 A0A336LZT5 A0A1S4H0Z5 W5JD82 A0A067RBY4 A0A182F5D2 A0A182KUK4 A0A2R7WXQ2 N6TXI0 A0A182V957 Q7PVZ6 A0A0A1XT83 A0A0K8WG59 A0A1W4V6X6 Q8T3J2 B4MWL0 B3MNI3 Q2XY32 P91643 Q9VK54 Q2XY29 Q2XY28 Q2XY30 Q2XY31 B4IEC7 B4P2J5 L7XBB4 A0A0J9R1M2 B4HCX9 Q2XY27 B3N3C1 B4JA35 B4LST1 Q29L68 A0A3B0KAM7 A0A2A3EKJ6 B4Q498 B4KGL2 A0A087ZPS3 A0A0M9A3L1 A0A310S8P8 A0A1S3D0I3 A0A1I8MH62 A0A0L0BX47 E2BHT5 A0A1A9Z7U9 A0A1A9V871 A0A1B0BAG3 E2AXW2 A0A1I8P7S1 A0A1A9XEN7 A0A1A9WXL5 A0A195EDT3 A0A195EUQ4 A0A151IP40 A0A158NXN6 A0A0J7KRU9 A0A3L8E423 A0A026WJP7 A0A2H8TQI4 A0A2S2NE99 J9K4V7 A0A154P0C2
Pubmed
19121390
26227816
22118469
26354079
28004739
18362917
+ More
19820115 17510324 23537049 29403074 24438588 25244985 26823975 25401762 12364791 20920257 23761445 24845553 20966253 25830018 17994087 16120803 8812109 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 15632085 25315136 26108605 20798317 21347285 30249741 24508170
19820115 17510324 23537049 29403074 24438588 25244985 26823975 25401762 12364791 20920257 23761445 24845553 20966253 25830018 17994087 16120803 8812109 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 15632085 25315136 26108605 20798317 21347285 30249741 24508170
EMBL
BABH01033382
ODYU01004612
SOQ44677.1
JTDY01006142
KOB66244.1
NWSH01002640
+ More
PCG67777.1 AGBW02012378 OWR45154.1 KQ459603 KPI92701.1 KQ460845 KPJ12086.1 GEZM01081350 JAV61695.1 AJVK01001761 DS232524 EDS43015.1 KQ971338 EFA02799.1 CH477216 EAT47531.1 GEDC01018218 JAS19080.1 KB632017 ERL88063.1 NEVH01001337 PNF43066.1 PYGN01000008 PSN58172.1 APGK01026716 KB740637 ENN79810.1 ATLV01022537 KE525333 KFB47751.1 CVRI01000001 CRK86405.1 APGK01002625 KB734878 ENN83491.1 ACPB03005451 APCN01003764 AXCM01017736 GDHC01013123 JAQ05506.1 GBHO01037691 JAG05913.1 UFQS01000368 UFQT01000368 SSX03230.1 SSX23596.1 AAAB01008984 ADMH02001719 ETN61313.1 KK852559 KDR21401.1 KK855900 PTY24317.1 APGK01009872 KB738060 ENN82793.1 EAA15120.4 GBXI01000016 JAD14276.1 GDHF01029256 GDHF01002280 JAI23058.1 JAI50034.1 AY095093 AAM11421.1 CH963857 EDW76151.1 CH902620 EDV32091.1 DQ138830 ABA86436.1 U42767 AAC47404.1 AE014134 AAF53225.3 ALI30206.1 DQ138833 ABA86439.1 DQ138834 ABA86440.1 DQ138832 ABA86438.1 DQ138831 ABA86437.1 CH480831 EDW45954.1 CM000157 EDW88225.2 BT149888 AGD79318.1 CM002910 KMY89978.1 CH479507 EDW33264.1 DQ138835 ABA86441.1 CH954177 EDV58761.1 CH916368 EDW03709.1 CH940649 EDW63820.1 CH379061 EAL32956.2 OUUW01000006 SPP82091.1 KZ288227 PBC31772.1 CM000361 EDX04848.1 CH933807 EDW13212.1 KQ435746 KOX76535.1 KQ769958 OAD52721.1 JRES01001205 KNC24628.1 GL448324 EFN84768.1 JXJN01010999 GL443736 EFN61726.1 KQ979074 KYN23004.1 KQ981958 KYN31990.1 KQ976914 KYN07108.1 ADTU01003250 LBMM01003877 KMQ93031.1 QOIP01000001 RLU27434.1 KK107168 EZA56183.1 GFXV01004662 MBW16467.1 GGMR01002932 MBY15551.1 ABLF02025983 KQ434782 KZC04590.1
PCG67777.1 AGBW02012378 OWR45154.1 KQ459603 KPI92701.1 KQ460845 KPJ12086.1 GEZM01081350 JAV61695.1 AJVK01001761 DS232524 EDS43015.1 KQ971338 EFA02799.1 CH477216 EAT47531.1 GEDC01018218 JAS19080.1 KB632017 ERL88063.1 NEVH01001337 PNF43066.1 PYGN01000008 PSN58172.1 APGK01026716 KB740637 ENN79810.1 ATLV01022537 KE525333 KFB47751.1 CVRI01000001 CRK86405.1 APGK01002625 KB734878 ENN83491.1 ACPB03005451 APCN01003764 AXCM01017736 GDHC01013123 JAQ05506.1 GBHO01037691 JAG05913.1 UFQS01000368 UFQT01000368 SSX03230.1 SSX23596.1 AAAB01008984 ADMH02001719 ETN61313.1 KK852559 KDR21401.1 KK855900 PTY24317.1 APGK01009872 KB738060 ENN82793.1 EAA15120.4 GBXI01000016 JAD14276.1 GDHF01029256 GDHF01002280 JAI23058.1 JAI50034.1 AY095093 AAM11421.1 CH963857 EDW76151.1 CH902620 EDV32091.1 DQ138830 ABA86436.1 U42767 AAC47404.1 AE014134 AAF53225.3 ALI30206.1 DQ138833 ABA86439.1 DQ138834 ABA86440.1 DQ138832 ABA86438.1 DQ138831 ABA86437.1 CH480831 EDW45954.1 CM000157 EDW88225.2 BT149888 AGD79318.1 CM002910 KMY89978.1 CH479507 EDW33264.1 DQ138835 ABA86441.1 CH954177 EDV58761.1 CH916368 EDW03709.1 CH940649 EDW63820.1 CH379061 EAL32956.2 OUUW01000006 SPP82091.1 KZ288227 PBC31772.1 CM000361 EDX04848.1 CH933807 EDW13212.1 KQ435746 KOX76535.1 KQ769958 OAD52721.1 JRES01001205 KNC24628.1 GL448324 EFN84768.1 JXJN01010999 GL443736 EFN61726.1 KQ979074 KYN23004.1 KQ981958 KYN31990.1 KQ976914 KYN07108.1 ADTU01003250 LBMM01003877 KMQ93031.1 QOIP01000001 RLU27434.1 KK107168 EZA56183.1 GFXV01004662 MBW16467.1 GGMR01002932 MBY15551.1 ABLF02025983 KQ434782 KZC04590.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000007151
UP000053268
UP000053240
+ More
UP000092462 UP000002320 UP000007266 UP000008820 UP000030742 UP000235965 UP000245037 UP000019118 UP000030765 UP000183832 UP000075880 UP000015103 UP000075840 UP000075902 UP000076407 UP000075901 UP000075900 UP000076408 UP000075885 UP000075884 UP000075920 UP000075883 UP000192223 UP000075881 UP000000673 UP000027135 UP000069272 UP000075882 UP000075903 UP000007062 UP000192221 UP000007798 UP000007801 UP000000803 UP000001292 UP000002282 UP000008744 UP000008711 UP000001070 UP000008792 UP000001819 UP000268350 UP000242457 UP000000304 UP000009192 UP000005203 UP000053105 UP000079169 UP000095301 UP000037069 UP000008237 UP000092445 UP000078200 UP000092460 UP000000311 UP000095300 UP000092443 UP000091820 UP000078492 UP000078541 UP000078542 UP000005205 UP000036403 UP000279307 UP000053097 UP000007819 UP000076502
UP000092462 UP000002320 UP000007266 UP000008820 UP000030742 UP000235965 UP000245037 UP000019118 UP000030765 UP000183832 UP000075880 UP000015103 UP000075840 UP000075902 UP000076407 UP000075901 UP000075900 UP000076408 UP000075885 UP000075884 UP000075920 UP000075883 UP000192223 UP000075881 UP000000673 UP000027135 UP000069272 UP000075882 UP000075903 UP000007062 UP000192221 UP000007798 UP000007801 UP000000803 UP000001292 UP000002282 UP000008744 UP000008711 UP000001070 UP000008792 UP000001819 UP000268350 UP000242457 UP000000304 UP000009192 UP000005203 UP000053105 UP000079169 UP000095301 UP000037069 UP000008237 UP000092445 UP000078200 UP000092460 UP000000311 UP000095300 UP000092443 UP000091820 UP000078492 UP000078541 UP000078542 UP000005205 UP000036403 UP000279307 UP000053097 UP000007819 UP000076502
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
H9JMA6
A0A2H1VV08
A0A0L7KT99
A0A2A4J6M8
A0A212EUM8
A0A194PH25
+ More
A0A194R895 A0A1Y1KPV8 A0A1B0GQT1 B0X983 D2A216 Q17LD1 A0A1S4EYJ9 A0A1B6D059 U4U6V4 A0A2J7RQF6 A0A2P8ZNT7 N6UDM9 A0A084WC07 A0A1J1HEI3 A0A182ITX6 N6URS7 T1HFN4 A0A182I527 A0A182U8F1 A0A182WYP4 A0A182TB69 A0A182R9K7 A0A182YCJ8 A0A182P6T4 A0A182NF07 A0A182WH30 A0A182MP10 A0A146LEI1 A0A1W4WR46 A0A0A9WHB9 A0A182KHJ8 A0A336LZT5 A0A1S4H0Z5 W5JD82 A0A067RBY4 A0A182F5D2 A0A182KUK4 A0A2R7WXQ2 N6TXI0 A0A182V957 Q7PVZ6 A0A0A1XT83 A0A0K8WG59 A0A1W4V6X6 Q8T3J2 B4MWL0 B3MNI3 Q2XY32 P91643 Q9VK54 Q2XY29 Q2XY28 Q2XY30 Q2XY31 B4IEC7 B4P2J5 L7XBB4 A0A0J9R1M2 B4HCX9 Q2XY27 B3N3C1 B4JA35 B4LST1 Q29L68 A0A3B0KAM7 A0A2A3EKJ6 B4Q498 B4KGL2 A0A087ZPS3 A0A0M9A3L1 A0A310S8P8 A0A1S3D0I3 A0A1I8MH62 A0A0L0BX47 E2BHT5 A0A1A9Z7U9 A0A1A9V871 A0A1B0BAG3 E2AXW2 A0A1I8P7S1 A0A1A9XEN7 A0A1A9WXL5 A0A195EDT3 A0A195EUQ4 A0A151IP40 A0A158NXN6 A0A0J7KRU9 A0A3L8E423 A0A026WJP7 A0A2H8TQI4 A0A2S2NE99 J9K4V7 A0A154P0C2
A0A194R895 A0A1Y1KPV8 A0A1B0GQT1 B0X983 D2A216 Q17LD1 A0A1S4EYJ9 A0A1B6D059 U4U6V4 A0A2J7RQF6 A0A2P8ZNT7 N6UDM9 A0A084WC07 A0A1J1HEI3 A0A182ITX6 N6URS7 T1HFN4 A0A182I527 A0A182U8F1 A0A182WYP4 A0A182TB69 A0A182R9K7 A0A182YCJ8 A0A182P6T4 A0A182NF07 A0A182WH30 A0A182MP10 A0A146LEI1 A0A1W4WR46 A0A0A9WHB9 A0A182KHJ8 A0A336LZT5 A0A1S4H0Z5 W5JD82 A0A067RBY4 A0A182F5D2 A0A182KUK4 A0A2R7WXQ2 N6TXI0 A0A182V957 Q7PVZ6 A0A0A1XT83 A0A0K8WG59 A0A1W4V6X6 Q8T3J2 B4MWL0 B3MNI3 Q2XY32 P91643 Q9VK54 Q2XY29 Q2XY28 Q2XY30 Q2XY31 B4IEC7 B4P2J5 L7XBB4 A0A0J9R1M2 B4HCX9 Q2XY27 B3N3C1 B4JA35 B4LST1 Q29L68 A0A3B0KAM7 A0A2A3EKJ6 B4Q498 B4KGL2 A0A087ZPS3 A0A0M9A3L1 A0A310S8P8 A0A1S3D0I3 A0A1I8MH62 A0A0L0BX47 E2BHT5 A0A1A9Z7U9 A0A1A9V871 A0A1B0BAG3 E2AXW2 A0A1I8P7S1 A0A1A9XEN7 A0A1A9WXL5 A0A195EDT3 A0A195EUQ4 A0A151IP40 A0A158NXN6 A0A0J7KRU9 A0A3L8E423 A0A026WJP7 A0A2H8TQI4 A0A2S2NE99 J9K4V7 A0A154P0C2
PDB
3ZYO
E-value=9.51325e-27,
Score=301
Ontologies
GO
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.999010
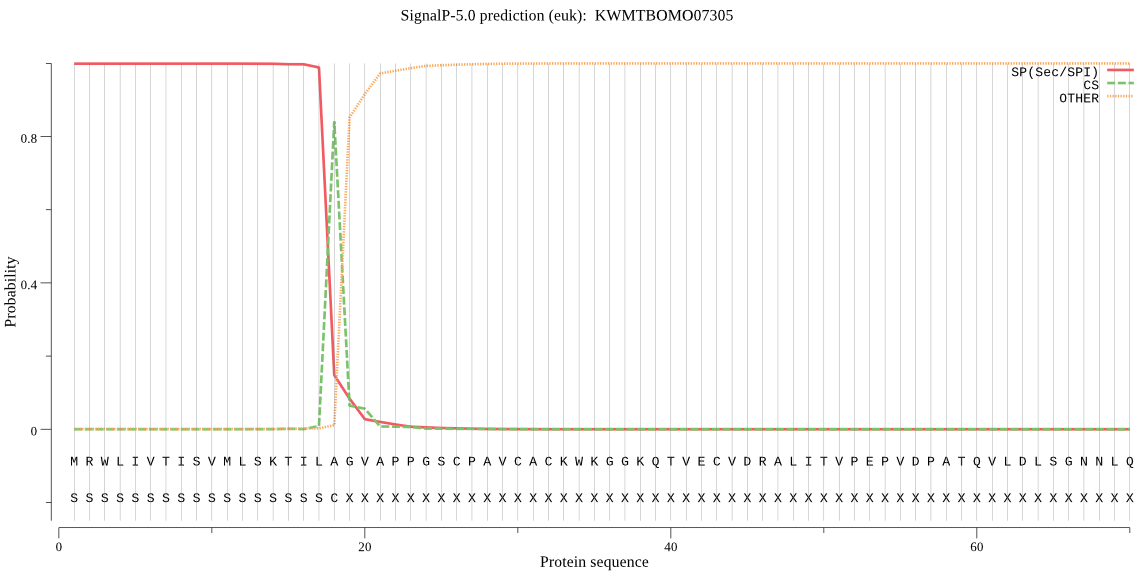
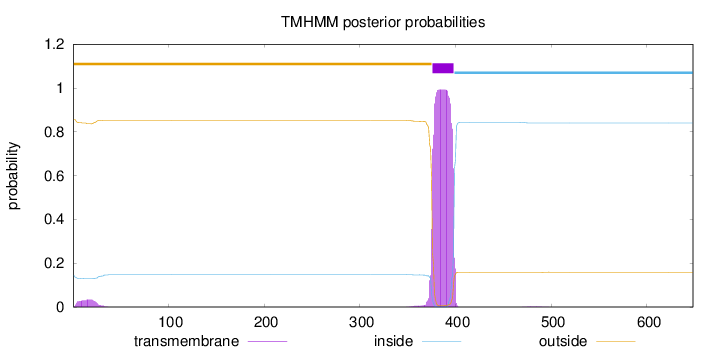
Length:
648
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.41695
Exp number, first 60 AAs:
0.73021
Total prob of N-in:
0.14532
outside
1 - 375
TMhelix
376 - 398
inside
399 - 648
Population Genetic Test Statistics
Pi
5.954429
Theta
6.829366
Tajima's D
-0.538486
CLR
13.022327
CSRT
0.242837858107095
Interpretation
Possibly Positive selection
